Ecosyste.ms: Awesome
An open API service indexing awesome lists of open source software.
https://github.com/kgorze/molecular-dynamics-simulator
A 2D molecular dynamics simulator that models atomic interactions using Lennard-Jones forces on a Boltzmann lattice. Currently implements an all-pairs algorithm for force calculations with visualization capabilities. This academic project aims to evolve into a 3D simulation of water and solutes.
https://github.com/kgorze/molecular-dynamics-simulator
atom-simulation cpp lennard-jones molecular-dynamics physics-simulation
Last synced: 3 days ago
JSON representation
A 2D molecular dynamics simulator that models atomic interactions using Lennard-Jones forces on a Boltzmann lattice. Currently implements an all-pairs algorithm for force calculations with visualization capabilities. This academic project aims to evolve into a 3D simulation of water and solutes.
- Host: GitHub
- URL: https://github.com/kgorze/molecular-dynamics-simulator
- Owner: kGorze
- Created: 2024-03-19T20:57:37.000Z (11 months ago)
- Default Branch: main
- Last Pushed: 2025-01-29T20:31:58.000Z (16 days ago)
- Last Synced: 2025-01-29T21:26:31.617Z (16 days ago)
- Topics: atom-simulation, cpp, lennard-jones, molecular-dynamics, physics-simulation
- Language: C++
- Homepage:
- Size: 160 MB
- Stars: 0
- Watchers: 1
- Forks: 0
- Open Issues: 24
-
Metadata Files:
- Readme: README.md
Awesome Lists containing this project
README
## Project Name
Molecular Dynamic Simulator - Simulates the molecular dynamics of atoms.
---
## Project Description
**This is an academic project focused on developing a molecular dynamics simulator.** Currently, it simulates Lennard-Jones (LJ) forces using a (less efficient) all-pairs algorithm on a 2D Boltzmann lattice of atoms. Half an assignment for a project in object-oriented programming and on the other hand a hobby project related to simulations. Lab assignment for Dr. Carrascoza - Bioinformatics 2nd semester, object-oriented programming.
**Am not familiar with Docker yet**, so here's a breakdown of the "toolchain" dependencies needed for the project:
Those are the files required to:
- run the main program, this is a zip archive with **Google test framework library** and **external libraries(inih, matplotplusplus).**
https://drive.google.com/file/d/1qWKXbvHN7cqKYxY8W0PSXjXOvH26tP9q/view?usp=sharing
- visualize the atom coordinates, this is a zip with **python scripts**
https://drive.google.com/file/d/1tgm48HFup8QAHkeGqLZC07GnXcTWbKDC/view?usp=sharing
### Initial Conditions:
Velocities and accelerations are **randomly assigned**, preventing the exact reproduction of simulations at present.
### Data Output:
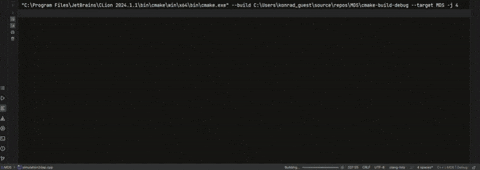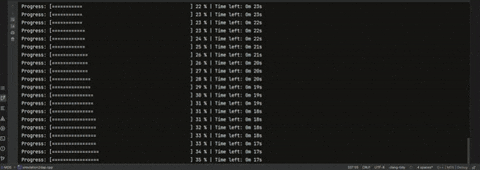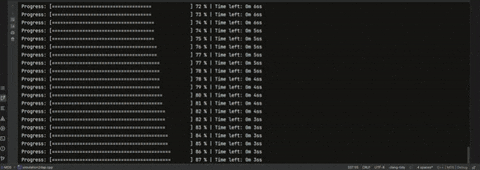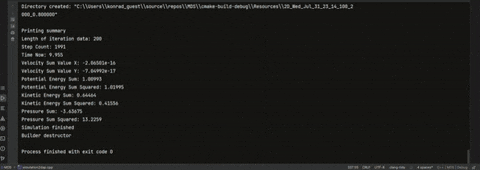
Each iteration is saved with **coordinates**, **physical properties**, and **velocity distribution** for later analysis.
### Visualization:
A separate **Python script** utilizing Pygame generates a visual representation of the atomic plane.
### Calculations:
### Visualization:
**Color coded** made by python script:
```
draw_atoms_velocity_sensitive.py
```
**No colour coded** by python script:
```
draw_atoms_no_color_coded.py
```
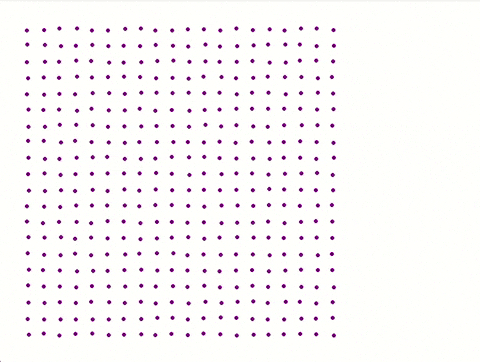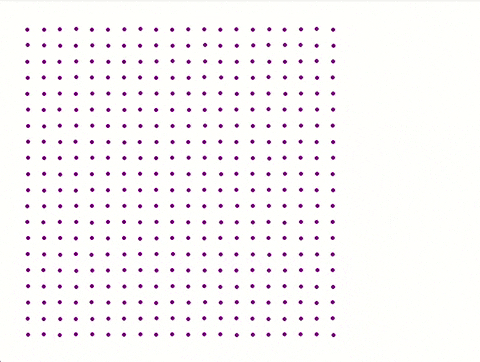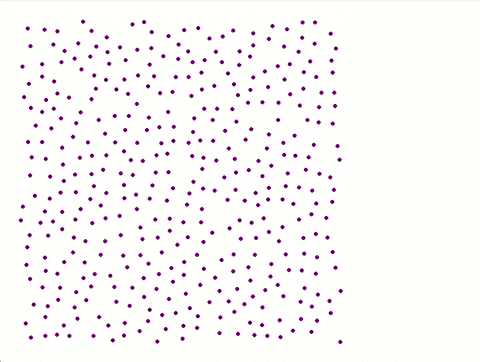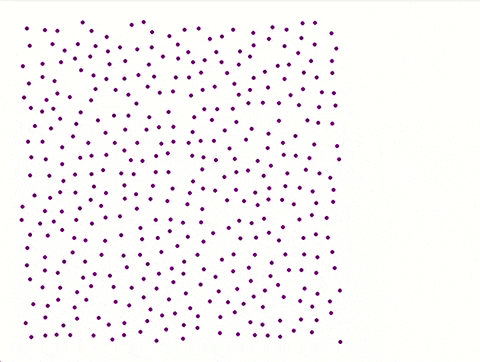
---
## Project Status
**This project is in its initial stages.** The ultimate goal is a 3D simulation of water and solutes, but the current focus is on achieving a stable 2D simulation of atoms.
### Stability:
The 2D simulation functionality is still under development.
---
## Features
The project employs a director-builder pattern to create a simulation object. The `computeForces` function calculates the forces acting on the atoms.
---
## Requirements
### For main program
#### Development Environment:
CLion
#### Compiler:
Likely GCC (**as used by CLion**)
### For python visualizations
pygame
---
## Installation
The program is not only intended for **Windows** environment and **JetBrains CLion IDE.**
[installation youtube instruction](https://www.youtube.com/watch?v=NE3JG-eTcnU)
---
## Usage Examples
The configuration file can be modified to obtain different simulation results.
---
## Documentation
Doxygen documentation is not yet available.
---
## Testing
Unit tests are still under development. The Google Test framework is being considered for implementation.
---
## Contribution and Issue Reporting
Feel free to fork the project or report issues for collaboration. No formal oversight exists yet.
---
## License
The code is freely reusable. The license for the book-based portions is still under consideration.
---
## Authors and Acknowledgments
This project is heavily inspired by the book: [The Art of Molecular Dynamics Simulation](https://www.cambridge.org/core/books/art-of-molecular-dynamics-simulation/57D40C5ECE9B7EA17C0E77E7754F5874)
1. Rapaport DC. _The Art of Molecular Dynamics Simulation_. 2nd ed. Cambridge University Press; 2004.
--
## Post-Mortem
First Iteration:
The initial challenge arose when we needed to create 3D simulations using methods originally designed for 2D. As a quick solution, we implemented a very primitive object-oriented paradigm with a deeply hierarchical structure.
Second Iteration:
The structure became an issue again when we had to incorporate design patterns for the final project submission. To address this, we rewrote the program, adding design patterns like Factory and Builder.
Third Iteration:
The third challenge was related to making the base class too generic, which hindered further development. To overcome this, we:
Created a UML diagram before coding.
Added design patterns to promote code reusability, modularity, and scalability.
Implemented a CI/DI framework with Google tests.
Divided the work into modules responsible for multithreading, inter-module communication, physics, etc.
Lessons Learned:
Early planning: Creating a UML diagram from the beginning can prevent architectural issues.
Design patterns: They are essential for maintainable and extensible code.
Testing: A comprehensive test suite is crucial for catching regressions.
Modularization: Breaking down the system into smaller, well-defined modules improves understanding and maintainability.