https://github.com/korniichuk/data-mining
Data Mining in R
https://github.com/korniichuk/data-mining
classification data-mining decision-tree jupyter jupyter-notebook machine-learning ml r r-language
Last synced: 4 months ago
JSON representation
Data Mining in R
- Host: GitHub
- URL: https://github.com/korniichuk/data-mining
- Owner: korniichuk
- License: unlicense
- Created: 2018-06-25T14:15:40.000Z (about 7 years ago)
- Default Branch: master
- Last Pushed: 2018-07-26T21:40:13.000Z (almost 7 years ago)
- Last Synced: 2025-01-17T08:28:24.867Z (6 months ago)
- Topics: classification, data-mining, decision-tree, jupyter, jupyter-notebook, machine-learning, ml, r, r-language
- Language: Jupyter Notebook
- Homepage: http://www.korniichuk.com
- Size: 1.53 MB
- Stars: 1
- Watchers: 2
- Forks: 0
- Open Issues: 0
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
README
# Data Mining with R
## Table of Contents
* **[Data Overview](#data-overview)**
* **[Summary Statistics](#summary-statistics)**
* **[Mean](#mean)**
* **[Median](#median)**
* **[All-in-One](#all-in-one)**
* **[Correlation](#correlation)**
* **[Graphics](#graphics)**
* **[Histograms](#histograms)**
* **[Boxplots](#boxplots)**
* **[Near Zero Variance Predictors](#near-zero-variance-predictors)**
* **[Linear Combinations](#linear-combinations)**
* **[Highly Correlated Variables](#highly-correlated-variables)**
* **[Distribution](#distribution)**
* **[Decision Tree](#decision-tree)**
* **[Classification](#classification)**
* **[SVM](#svm)**
* **[KNN](#knn)**
* **[SVM vs KNN](#svm-vs-knn)**
Import libs:
```R
library(caret)
library(data.table)
library(dplyr)
library(PerformanceAnalytics)
library(rpart.plot)
```
## Data Overview
Data Set Characteristics: | Number of Instances: | Attribute Characteristics: | Number of Attributes: | Associated Tasks:
--- | --- | --- | --- | ---
Multivariate | 1372| Real | 5| Classification
**Dataset information:** Data were extracted from images that were taken from genuine and forged banknote-like specimens. For digitization, an industrial camera usually used for print inspection was used. The final images have 400 x 400 pixels. Due to the object lens and distance to the investigated object gray-scale pictures with a resolution of about 660 dpi were gained. Wavelet Transform tool were used to extract features from images.
**Attribute information:**
1. variance of Wavelet Transformed image (type: continuous)
2. skewness of Wavelet Transformed image (type: continuous)
3. curtosis of Wavelet Transformed image (type: continuous)
4. entropy of image (type: continuous)
5. class (type: integer)
**Data source:** https://archive.ics.uci.edu/ml/datasets/banknote+authentication
Load `data_banknote_authentication.txt` file:
```R
url = paste('https://archive.ics.uci.edu/ml/machine-learning-databases/00267/',
'data_banknote_authentication.txt', sep='')
df = data.frame(fread(url))
names(df) = c('variance', 'skewness', 'curtosis', 'entropy', 'class')
```
Check size of `df` dataframe:
```R
nrow(df)
```
**Output**:
```
1372
```
Show the first part of `df` dataframe:
```R
head(df, 5)
```
varianceskewnesscurtosisentropyclass
3.62160 8.6661 -2.8073 -0.446990
4.54590 8.1674 -2.4586 -1.462100
3.86600 -2.6383 1.9242 0.106450
3.45660 9.5228 -4.0112 -3.594400
0.32924 -4.4552 4.5718 -0.988800
Show the last part of `df` dataframe:
```R
tail(df, 5)
```
varianceskewnesscurtosisentropyclass
1368 0.40614 1.34920-1.4501 -0.55949 1
1369-1.38870 -4.87730 6.4774 0.34179 1
1370-3.75030 -13.4586017.5932 -2.77710 1
1371-3.56370 -8.3827012.3930 -1.28230 1
1372-2.54190 -0.65804 2.6842 1.19520 1
## Summary Statistics
### Mean
```R
print(noquote(paste0('Mean. Variance of Wavelet Transformed image: ', mean(df$variance))))
print(noquote(paste0('Mean. Skewness of Wavelet Transformed image: ', mean(df$skewness))))
print(noquote(paste0('Mean. Curtosis of Wavelet Transformed image: ', mean(df$curtosis))))
print(noquote(paste0('Mean. Entropy of image: ', mean(df$entropy))))
```
**Output**:
```
[1] Mean. Variance of Wavelet Transformed image: 0.433735257069971
[1] Mean. Skewness of Wavelet Transformed image: 1.92235312063936
[1] Mean. Curtosis of Wavelet Transformed image: 1.39762711726676
[1] Mean. Entropy of image: -1.19165652004373
```
### Median
```R
print(noquote(paste0('Median. Variance of Wavelet Transformed image: ',
median(df$variance))))
print(noquote(paste0('Median. Skewness of Wavelet Transformed image: ',
median(df$skewness))))
print(noquote(paste0('Median. Curtosis of Wavelet Transformed image: ',
median(df$curtosis))))
print(noquote(paste0('Median. Entropy of image: ', median(df$entropy))))
```
**Output**:
```
[1] Median. Variance of Wavelet Transformed image: 0.49618
[1] Median. Skewness of Wavelet Transformed image: 2.31965
[1] Median. Curtosis of Wavelet Transformed image: 0.61663
[1] Median. Entropy of image: -0.58665
```
### All-in-One
```R
print(noquote('Summary:'))
summary(select(df, -class))
```
**Output**:
```
[1] Summary:
variance skewness curtosis entropy
Min. :-7.0421 Min. :-13.773 Min. :-5.2861 Min. :-8.5482
1st Qu.:-1.7730 1st Qu.: -1.708 1st Qu.:-1.5750 1st Qu.:-2.4135
Median : 0.4962 Median : 2.320 Median : 0.6166 Median :-0.5867
Mean : 0.4337 Mean : 1.922 Mean : 1.3976 Mean :-1.1917
3rd Qu.: 2.8215 3rd Qu.: 6.815 3rd Qu.: 3.1793 3rd Qu.: 0.3948
Max. : 6.8248 Max. : 12.952 Max. :17.9274 Max. : 2.4495
```
### Correlation
```R
cor(df)
```
varianceskewnesscurtosisentropyclass
variance 1.0000000 0.2640255 -0.3808500 0.27681670-0.72484314
skewness 0.2640255 1.0000000 -0.7868952 -0.52632084-0.44468776
curtosis-0.3808500 -0.7868952 1.0000000 0.31884089 0.15588324
entropy 0.2768167 -0.5263208 0.3188409 1.00000000-0.02342368
class-0.7248431 -0.4446878 0.1558832 -0.02342368 1.00000000
```R
chart.Correlation(select(df, -class), histogram=TRUE)
```

## Graphics
### Histograms
```R
par(mfrow=c(2,2))
hist(df$variance, main='Histogram of Variance',
xlab='Variance of Wavelet Transformed Image')
hist(df$skewness, main='Histogram of Skewness',
xlab='Skewness of Wavelet Transformed Image')
hist(df$curtosis, main='Histogram of Curtosis',
xlab='Curtosis of Wavelet Transformed Image')
hist(df$entropy, main='Histogram of Entropy',
xlab='Entropy of Image')
```

### Boxplots
```R
par(mfrow=c(2,2))
boxplot(df$variance, data=df, main='Boxplot. Variance', horizontal=TRUE)
boxplot(df$skewness, data=df, main='Boxplot. Skewness', horizontal=TRUE)
boxplot(df$curtosis, data=df, main='Boxplot. Curtosis', horizontal=TRUE)
boxplot(df$entropy, data=df, main='Boxplot. Entropy', horizontal=TRUE)
```

## Near Zero Variance Predictors
```R
nearZeroVar(select(df, -class), saveMetrics=TRUE)
```
freqRatiopercentUniquezeroVarnzv
variance1.25 97.52187FALSE FALSE
skewness1.20 91.54519FALSE FALSE
curtosis1.00 92.56560FALSE FALSE
entropy1.00 84.25656FALSE FALSE
## Linear Combinations
```R
findLinearCombos(select(df, -class))
```
**Output**:
```
$linearCombos
list()
$remove
NULL
```
## Highly Correlated Variables
```R
df$class = as.character(ifelse(df$class=='1', 'Y', 'N'))
df2 = select(df, -class)
cor_matrix = cor(df2)
print(noquote('Highly correlated variables:'))
summary(cor_matrix[upper.tri(cor_matrix)]) # upper triangular part of a matrix
```
**Output**:
```
[1] Highly correlated variables:
Min. 1st Qu. Median Mean 3rd Qu. Max.
-0.78690 -0.48995 -0.05841 -0.13906 0.27362 0.31884
```
```R
high_cor_var = findCorrelation(cor_matrix, cutoff = 0.75) # check var above 0.75
print(noquote(paste0('Highly correlated variables: ', names(df2)[high_cor_var])))
```
**Output**:
```
[1] Highly correlated variables: skewness
```
Delete highly correlated `skewness` column from dataframe:
```R
df2 = select(df2, -skewness)
```
```R
cor_matrix = cor(df2)
print(noquote('Highly correlated variables:'))
summary(cor_matrix[upper.tri(cor_matrix)]) # upper triangular part of a matrix
df = cbind.data.frame(df2, class = df$class) # add class
```
**Output**:
```
[1] Highly correlated variables:
Min. 1st Qu. Median Mean 3rd Qu. Max.
-0.38085 -0.05202 0.27682 0.07160 0.29783 0.31884
```
## Distribution
```R
print(noquote('Distribution:'))
table(df$class)
```
**Output**:
```
[1] Distribution:
N Y
762 610
```
```R
class_freq = data.frame(table(df$class))
names(class_freq) = c('class', 'freq')
percent_chart = cbind(class_freq,
percent=round((class_freq$freq/sum(class_freq$freq))*100, 1))
percent_chart
```
classfreqpercent
N 762 55.5
Y 610 44.5
```R
slices = percent_chart$percent
lbls = c('N', 'Y')
pct = round(slices/sum(slices)*100, 1)
lbls = paste(lbls, pct) # add values of pct to labels
lbls = paste(lbls, '%', sep='') # add % char to labels
pie(slices, labels=lbls, radius=1, main='Pie Chart of Distribution',
clockwise=TRUE)
```

```R
featurePlot(x=select(df, -class), y=df$class, plot='box')
```

## Decision Tree
```R
rtree_set = rpart(class ~ ., df)
prp(rtree_set)
```

## Classification
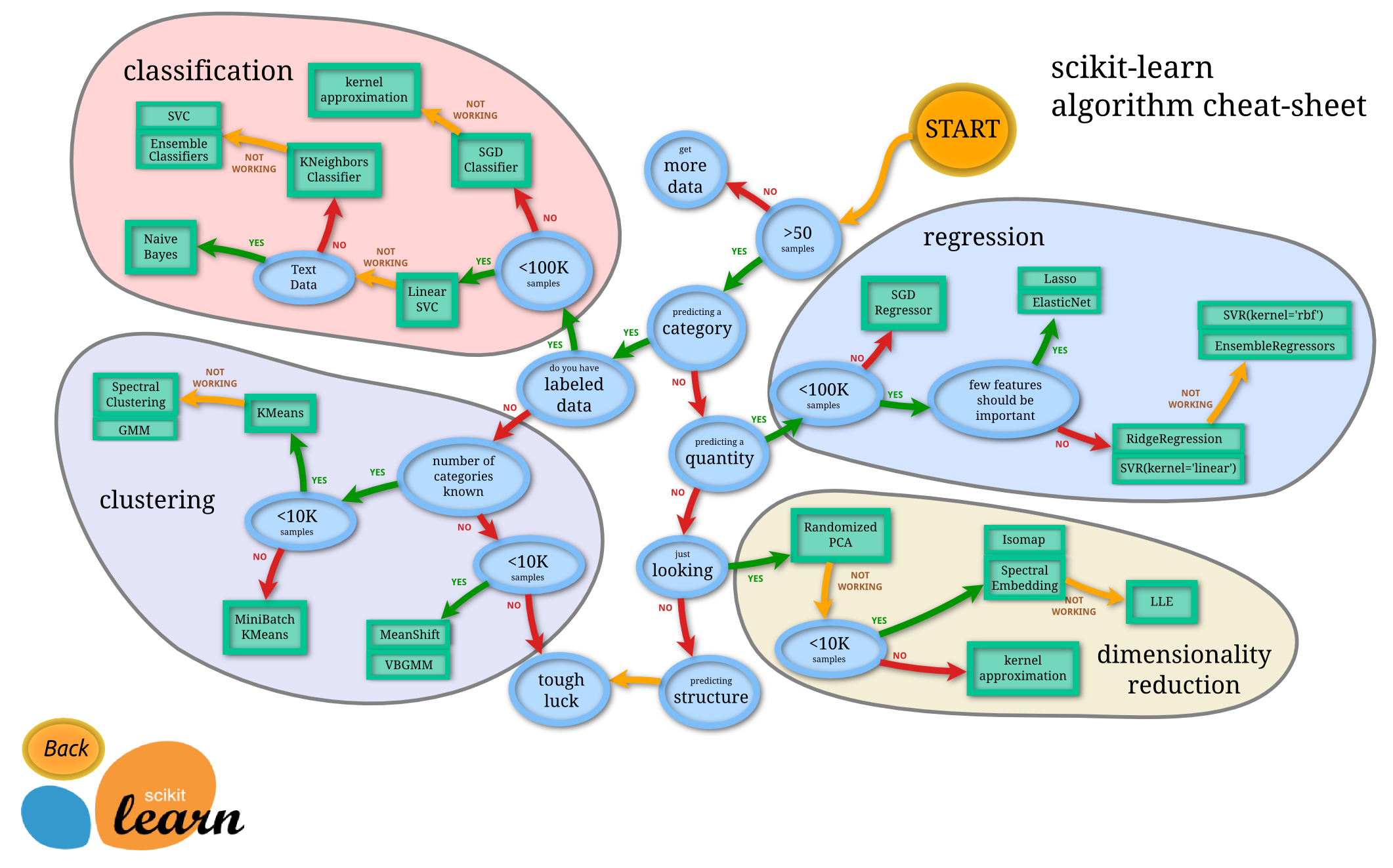
Split the data to train and test sets:
```R
train_ind = createDataPartition(df$class, p=0.7, list=FALSE)
data_train = data.frame(df[train_ind, ])
data_test = data.frame(df[-train_ind, ])
print(noquote('Train:'))
table(data_train$class)
print(noquote('Test:'))
table(data_test$class)
```
**Output**:
```
[1] Train:
N Y
534 427
[1] Test:
N Y
228 183
```
Choose validation method for the test of model:
```R
valid_par = trainControl(method='repeatedcv', number=5, repeats=10, p=0.70, preProc='range')
```
### SVM
```R
mod_svm = train(class ~ ., data=data_train, trControl=valid_par, method='svmRadial')
mod_svm
```
**Output**:
```
Support Vector Machines with Radial Basis Function Kernel
961 samples
3 predictors
2 classes: 'N', 'Y'
No pre-processing
Resampling: Cross-Validated (5 fold, repeated 10 times)
Summary of sample sizes: 768, 769, 770, 769, 768, 770, ...
Resampling results across tuning parameters:
C Accuracy Kappa
0.25 0.9785620 0.9568214
0.50 0.9807452 0.9612074
1.00 0.9815764 0.9628818
Tuning parameter 'sigma' was held constant at a value of 0.8345372
Accuracy was used to select the optimal model using the largest value.
The final values used for the model were sigma = 0.8345372 and C = 1.
```
### KNN
```R
mod_knn = train(class ~. , data=data_train, trControl=valid_par, method='knn')
mod_knn
```
**Output**:
```
k-Nearest Neighbors
961 samples
3 predictors
2 classes: 'N', 'Y'
No pre-processing
Resampling: Cross-Validated (5 fold, repeated 10 times)
Summary of sample sizes: 768, 769, 769, 769, 769, 768, ...
Resampling results across tuning parameters:
k Accuracy Kappa
5 0.9761700 0.9519131
7 0.9766871 0.9529962
9 0.9748131 0.9492504
Accuracy was used to select the optimal model using the largest value.
The final value used for the model was k = 7.
```
Show summary:
```R
print(noquote('Summary:'))
mod_results = resamples(list(SVM=mod_svm, KNN=mod_knn))
summary(mod_results)
```
**Output**:
```
[1] Summary:
Call:
summary.resamples(object = mod_results)
Models: SVM, KNN
Number of resamples: 50
Accuracy
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
SVM 0.9633508 0.9740933 0.9843750 0.9815764 0.9882606 1.0000000 0
KNN 0.9581152 0.9687905 0.9740933 0.9766871 0.9843750 0.9947917 0
Kappa
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
SVM 0.9264050 0.9478632 0.9684487 0.9628818 0.9762997 1.0000000 0
KNN 0.9157941 0.9372235 0.9476255 0.9529962 0.9684487 0.9894575 0
```
### SVM vs KNN
```R
bwplot(mod_results, scales=list(x=list(relation='free'), y=list(relation='free')))
```

Test models:
```R
test = select(data_test, -class)
test_sum = data_test$class
mod_predict_svm = predict(mod_svm, test)
print(noquote('SMV:'))
confusionMatrix(mod_predict_svm, test_sum)
mod_predict_knn = predict(mod_knn, test)
print(noquote('KNN:'))
confusionMatrix(mod_predict_knn, test_sum)
```
**Output**:
```
[1] SMV:
Confusion Matrix and Statistics
Reference
Prediction N Y
N 222 1
Y 6 182
Accuracy : 0.983
95% CI : (0.9652, 0.9931)
No Information Rate : 0.5547
P-Value [Acc > NIR] : <2e-16
Kappa : 0.9656
Mcnemar's Test P-Value : 0.1306
Sensitivity : 0.9737
Specificity : 0.9945
Pos Pred Value : 0.9955
Neg Pred Value : 0.9681
Prevalence : 0.5547
Detection Rate : 0.5401
Detection Prevalence : 0.5426
Balanced Accuracy : 0.9841
'Positive' Class : N
[1] KNN:
Confusion Matrix and Statistics
Reference
Prediction N Y
N 223 2
Y 5 181
Accuracy : 0.983
95% CI : (0.9652, 0.9931)
No Information Rate : 0.5547
P-Value [Acc > NIR] : <2e-16
Kappa : 0.9656
Mcnemar's Test P-Value : 0.4497
Sensitivity : 0.9781
Specificity : 0.9891
Pos Pred Value : 0.9911
Neg Pred Value : 0.9731
Prevalence : 0.5547
Detection Rate : 0.5426
Detection Prevalence : 0.5474
Balanced Accuracy : 0.9836
'Positive' Class : N
```