https://github.com/labsquare/cutepeaks
CutePeaks is a standalone Sanger trace viewer steered by a modern and user-friendly UI.
https://github.com/labsquare/cutepeaks
ab1 sanger-chromatograms scf
Last synced: 6 months ago
JSON representation
CutePeaks is a standalone Sanger trace viewer steered by a modern and user-friendly UI.
- Host: GitHub
- URL: https://github.com/labsquare/cutepeaks
- Owner: labsquare
- License: gpl-3.0
- Created: 2017-01-18T15:17:55.000Z (over 8 years ago)
- Default Branch: master
- Last Pushed: 2022-12-20T08:27:39.000Z (over 2 years ago)
- Last Synced: 2024-04-14T05:56:44.251Z (about 1 year ago)
- Topics: ab1, sanger-chromatograms, scf
- Language: C++
- Homepage: https://labsquare.github.io/CutePeaks/
- Size: 9.49 MB
- Stars: 38
- Watchers: 12
- Forks: 14
- Open Issues: 28
-
Metadata Files:
- Readme: README.md
- Changelog: CHANGELOG
- Contributing: CONTRIBUTING.md
- Funding: .github/FUNDING.yml
- License: LICENSE
- Code of conduct: CODE_OF_CONDUCT.md
- Citation: CITATION.cff
Awesome Lists containing this project
README
# CutePeaks
[](https://github.com/labsquare/CutePeaks/actions/workflows/c-cpp.yml)
[](https://doi.org/10.5281/zenodo.5148809)
[](https://doi.org/10.21105/joss.03457)
https://labsquare.github.io/CutePeaks/
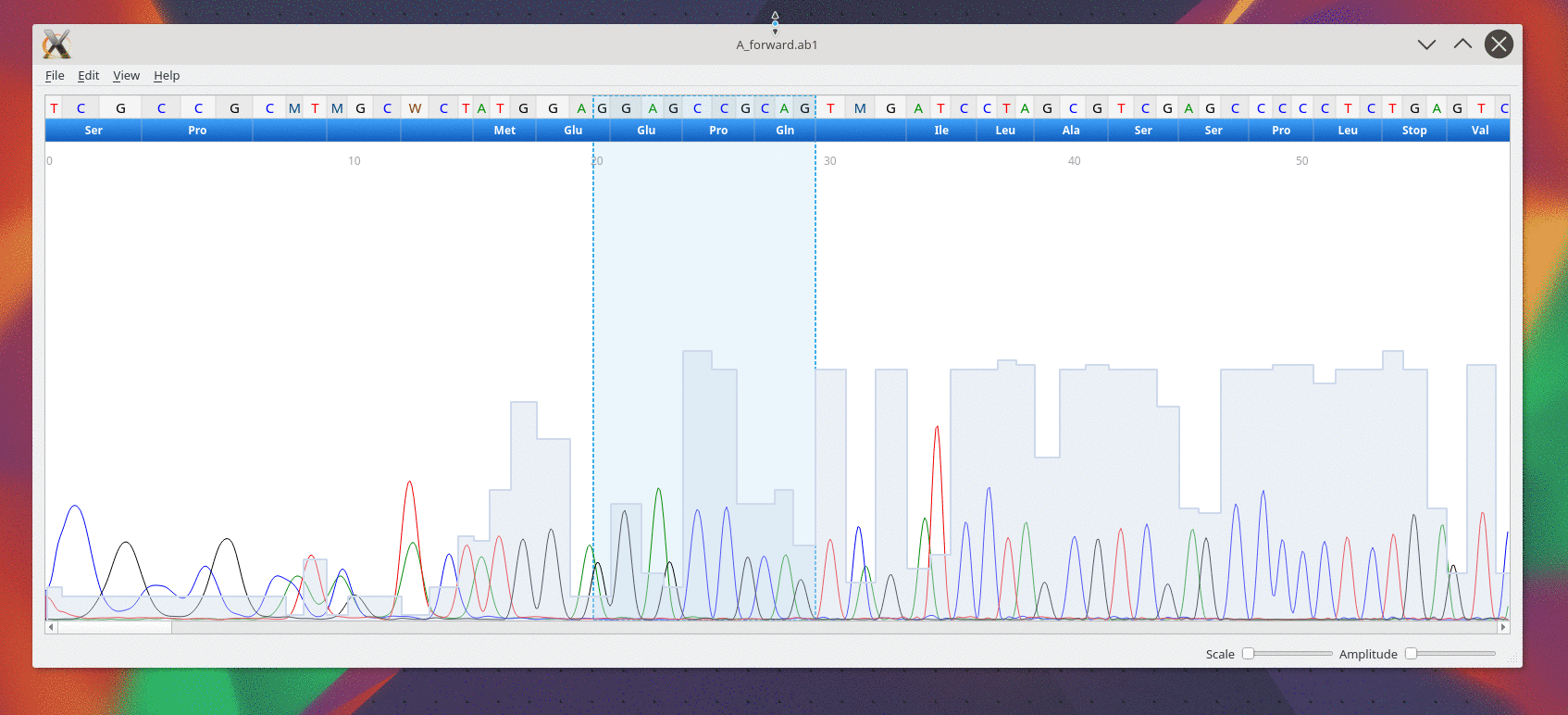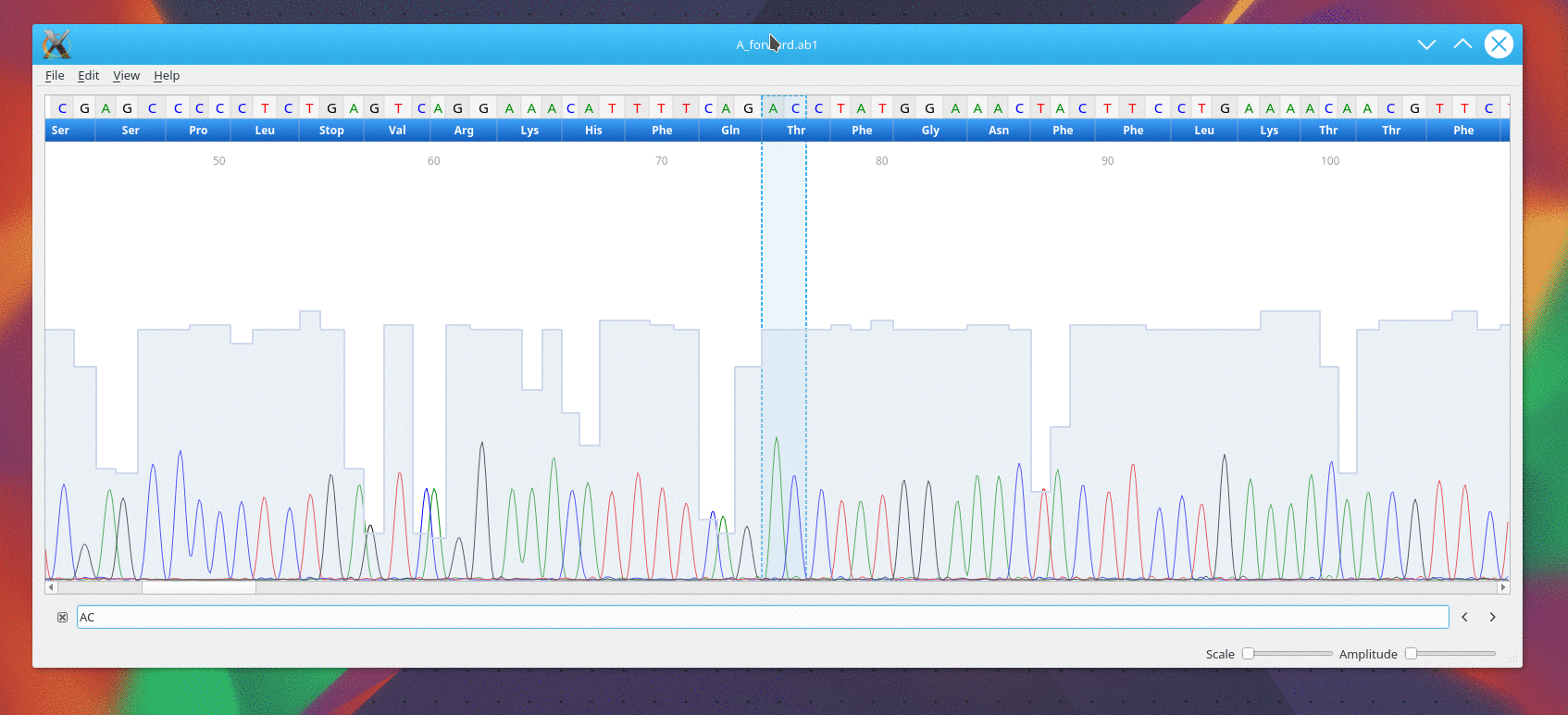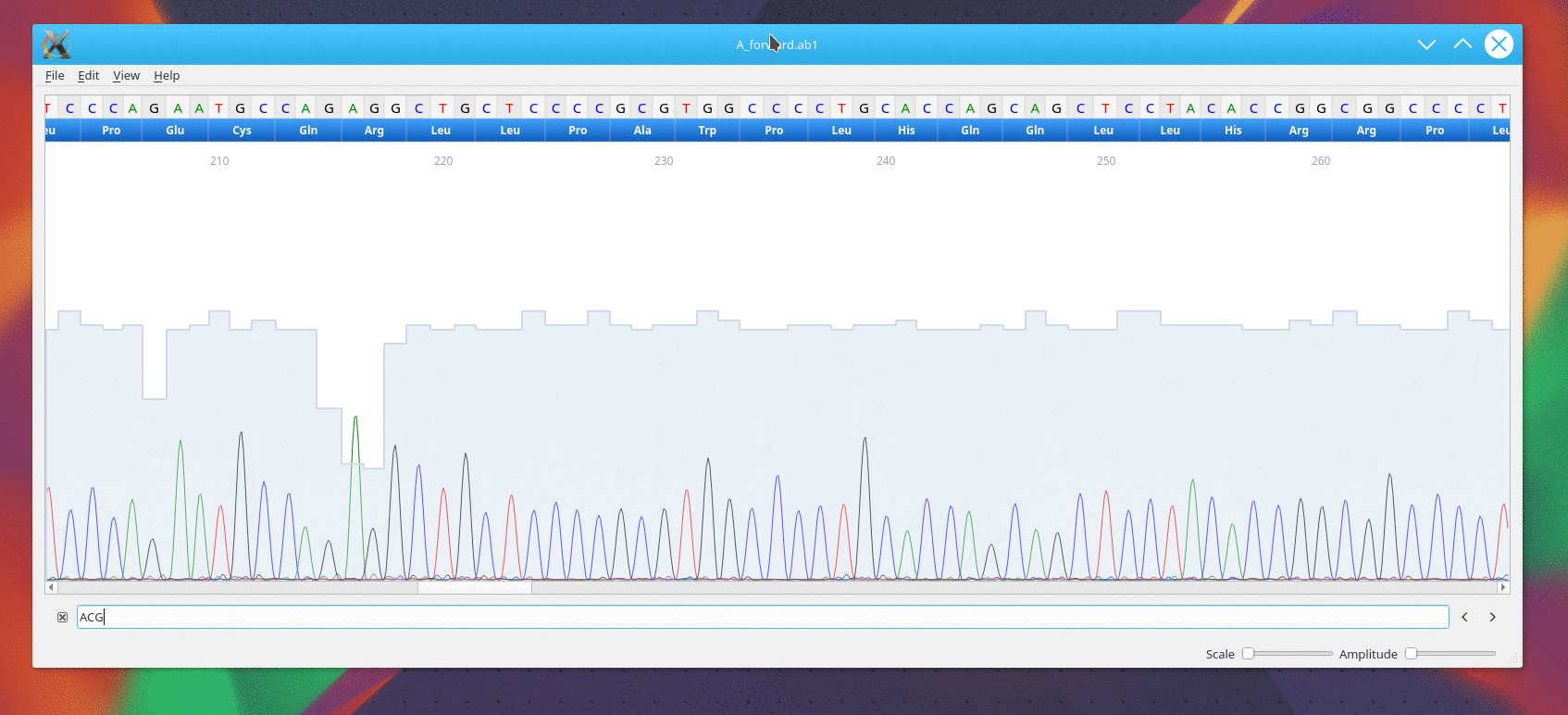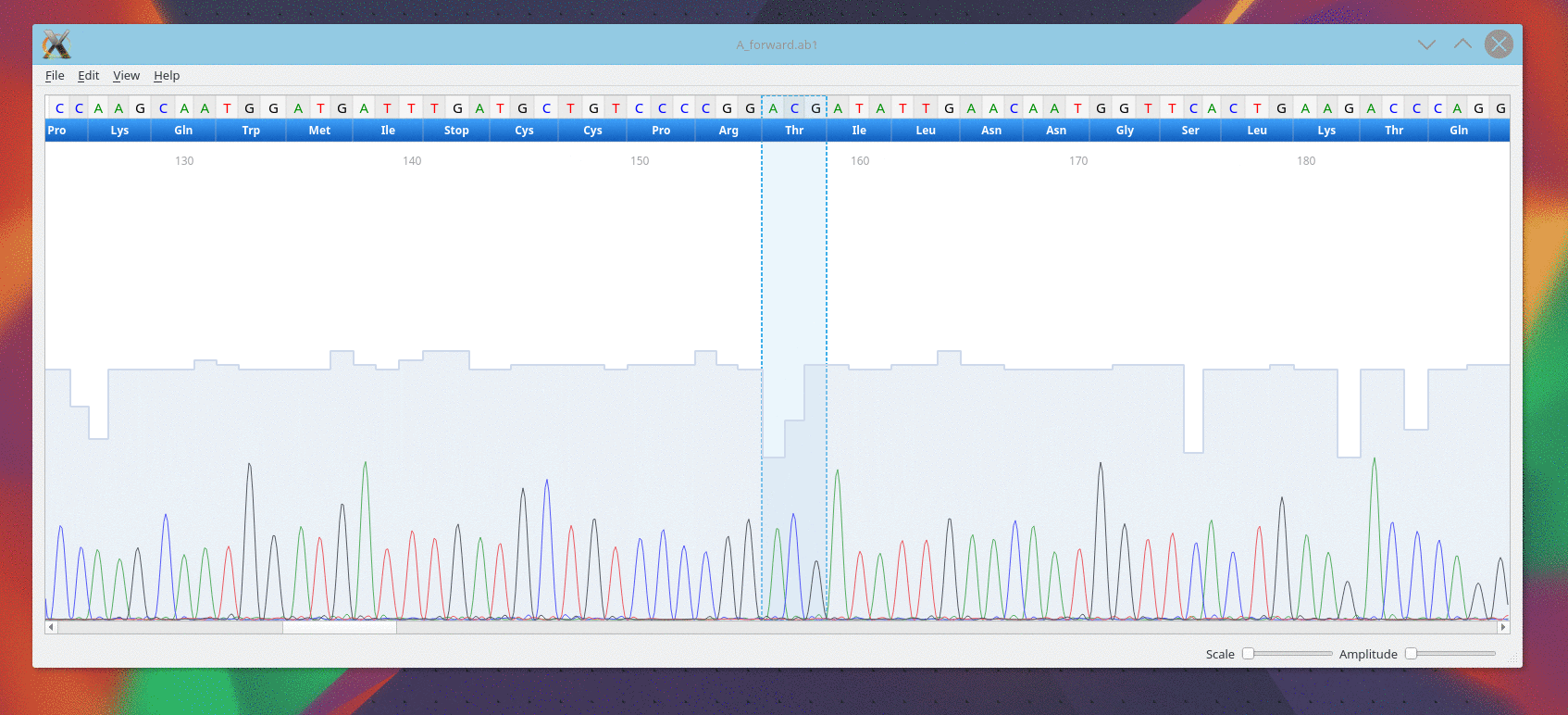
A simple viewer for Sanger trace file made with Qt5.
Supports AB1 and SCF 3.0 file formats.
It has regular expression pattern finder and can export trace as SVG vector image.
# Statement of need
Despite the major use of Next Generation Sequencing, the Sanger method is still widely used in genetic labs as the gold standard to read target DNA sequences. Very few opensource software is available to explore Sanger trace data and most of labs staff still rely on proprietary software. Moreover, they are not always user-friendly and lack modern look and feel.
# State of field
[4peaks](https://nucleobytes.com/4peaks/) is a software widely used by biologists that benefits from a nice User interface. Sadly, it is only available on MacOS and source code is not opened to community enhancement. [Seqtrace](https://github.com/stuckyb/seqtrace) is the only standalone and opensource application we could find. However, it is written with GTK framework in Python 2, the latter being deprecated and slower than C++.
# Installation
## Windows
[Download windows binary](https://github.com/labsquare/CutePeaks/releases/download/0.2.3/CutePeaks-win32.exe)
## MacOSX
[Downlad MacOS binary](https://github.com/labsquare/CutePeaks/releases/download/0.2.3/cutepeaks-macos.dmg)
## Linux
### AppImage
Linux binary is available as [AppImage](http://appimage.org/).
Download the AppImage from [here](https://github.com/labsquare/CutePeaks/releases).
For ubuntu >=21.04, Download this one [here](https://github.com/labsquare/CutePeaks/releases/download/0.2.3/cutepeaks-ubuntu_21-04-x86_64.AppImage)
Run it as follow:
chmod +x cutepeaks-0.2.0-linux-x86_64.appimage
./cutepeaks-0.2.0-linux-x86_64.appimage
### FlatPak
Cutepeaks is available as a [flatpak](https://flathub.org/apps/details/io.github.labsquare.CutePeaks)
```
flatpak install flathub io.github.labsquare.CutePeaks
flatpak run io.github.labsquare.CutePeaks
```
## Compilation
### Prerequisites
#### Install Qt ≥ 5.7
**From website**: Download Qt ≥ 5.7 from https://www.qt.io/.
Don't forget to check QtChart module during installation.
**From Ubuntu**: Qt 5.7 is not yet available with Ubuntu. But you can add a PPA to your software system.
For exemple for Xenial:
sudo add-apt-repository ppa:beineri/opt-qt57-xenial
sudo apt install qt57base qt57charts-no-lgpl
source /opt/qt57/bin/qt57-env.sh
**From Fedora**: Qt 5.7 is available.
sudo dnf install qt5-qtbase-devel qt5-qtcharts-devel
### Compile CutePeaks
Be sure you have the correct version of Qt (≥ 5.7) by using qmake --version. For exemple, if you have installed Qt from ppa:beineri, you will find it under /opt/qt57/bin/qmake. Then launch the compilation from CutePeaks folder as follow.
/opt/qt57/bin/qmake --version
/opt/qt57/bin/qmake
make
sudo make install
## Usage
CutePeaks supports following trace file formats:
- *.ab1
- *.scf
Example files are available here:
https://github.com/labsquare/CutePeaks/tree/master/examples
You can open those files from cutepeaks by clicking on *open* from the File menu.
## Features
Once the file is open, cutepeaks allows you to :
- Explore the trace from a scroll area. ( Finger geasture are supported with touch screen)
- Scale the trace horizontally or vertically using 2 sliders at the bottom right.
- Select a subsequence with the mouse as with any text editor. Then you can cut or copy to the clipboard
- Make the reverse complement from the edit menu
- Display Sequence and metadata from the view menu
- Search for a regular expression pattern. Open the "Find Sequence..." from the edit menu
- Export trace or sequence to different format. ( e.g: Fasta, CSV, SVG or PNG image )
## Contributions / Bugs
See [how to contributing](https://github.com/labsquare/CutePeaks/edit/master/CONTRIBUTING.md)
## Licenses
This program is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with this program. If not, see https://www.gnu.org/licenses/gpl-3.0.txt.