https://github.com/liubenyuan/pyEIT
Python based toolkit for Electrical Impedance Tomography
https://github.com/liubenyuan/pyEIT
Last synced: 3 months ago
JSON representation
Python based toolkit for Electrical Impedance Tomography
- Host: GitHub
- URL: https://github.com/liubenyuan/pyEIT
- Owner: liubenyuan
- License: other
- Fork: true (eitcom/pyEIT)
- Created: 2022-09-23T01:54:34.000Z (almost 3 years ago)
- Default Branch: master
- Last Pushed: 2023-10-24T13:14:30.000Z (over 1 year ago)
- Last Synced: 2025-02-09T09:30:06.538Z (5 months ago)
- Language: Python
- Homepage:
- Size: 1010 KB
- Stars: 6
- Watchers: 2
- Forks: 1
- Open Issues: 0
-
Metadata Files:
- Readme: README.md
- License: LICENSE.txt
Awesome Lists containing this project
- awesome-eit - pyEIT - based, open-source framework for Electrical Impedance Tomography (EIT). From **pyeit**. (EIT simulation libraries)
README
# 
**Note, the upstream of `pyeit` goes here: [eitcom/pyeit](https://github.com/eitcom/pyEIT).**
`pyEIT` is a python-based, open-source framework for Electrical Impedance Tomography (EIT). The design priciples of `pyEIT` are **modularity, minimalism, extensibility and OOP.**
## 1. Installation
`pyEIT` is purely python based, it can be installed and run without any difficulty.
### 1.1 Install using pip or conda-forge (recommended)
`pyEIT` is now availbale on `pypi` and `conda-forge`. It is purely python, which can be installed via
```bash
$ pip install pyeit
```
or
```bash
$ conda install -c conda-forge pyeit
```
### 1.2 Install from source code
You can track the git version of `pyEIT`, and use it locally by setting the `PYTHONPATH` variable.
```bash
export PYTHONPATH=/path/to/pyEIT
```
If you are using `spyder`, or `pyCharm`, you can also set `PYTHONPATH` per project in the IDE, which is more convenient.
Alternatively, but not recommended, you can compile and install from source code,
```bash
$ python setup.py build
$ python setup.py install
```
## 2. Run the examples
From the example folder, pick one demo and run!
**Note:** the following images may be outdated due to that the parameters of a EIT algorithm may be changed in different versions of `pyEIT`. And it is there in the code, so just run the demo.
### 2.1 (2D) forward and inverse computing
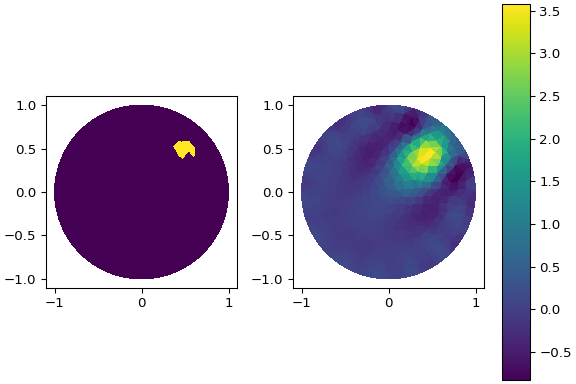
**Using** `examples/eit_dynamic_bp.py`
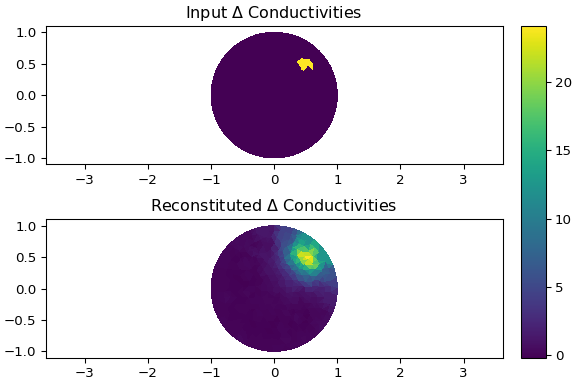
**Using** `examples/eit_dynamic_greit.py`
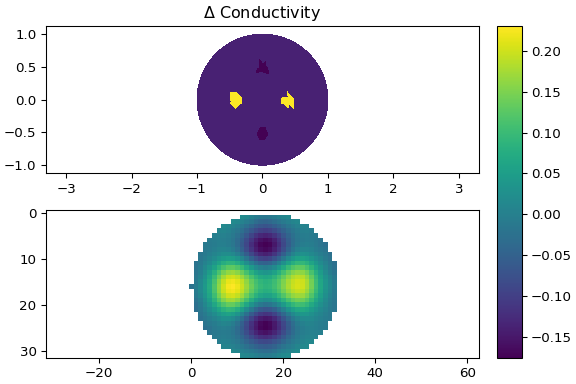
**Using** `examples/eit_dynamic_jac.py`
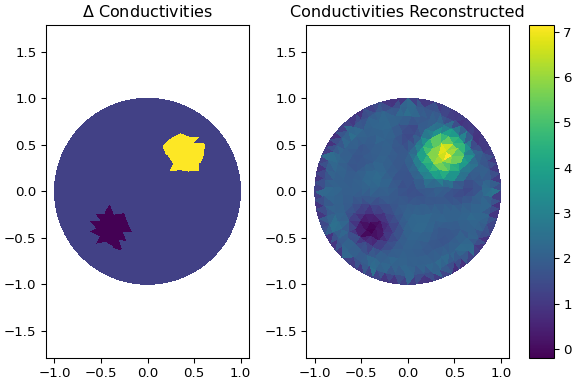
**Using** `examples/eit_static_jac.py`
### 2.2 (3D) forward and inverse computing
**Using** `examples/demo_forward3d.py`
**Using** `examples/demo_dynamic_jac3d.py`
**Limitations:**
1. 3D visualization plotted using `vispy` can be adjusted using mouse wheels interactively. Seeking a perfect visualization mode, transparency or opaque, is in fact an individual taste. User can also try `mayavi` and `vtk` for the visualization purpose using the unified 3D meshing structure.
2. Solving the inverse problem of 3D EIT, requires the electrodes to be placed at multiple altitude (z-axis) in order to have a (better) z-resolution. This should be done carefully, as adding extra z-belt introduces more stimulation patterns, which in turn adds to the computational loads.
## 3. Contribute to pyEIT
Give `pyEIT` a **star**, **fork** this project and commit a pull request **(PR)** !
### 3.1 Feature lists
- [x] 2D forward and inverse computing of EIT
- [x] Reconstruction algorithms : Gauss-Newton solver (JAC), Back-projection (BP), 2D GREIT
- [x] 2D/3D visualization!
- [x] Add support for 3D forward and inverse computing
- [x] 3D mesh generation and visualization
- [ ] Generate 2D/3D meshes from CT/MRI (based on iso2mesh [https://github.com/fangq/iso2mesh](https://github.com/fangq/iso2mesh))
- [ ] Complete electrode model (CEM) support
- [ ] Implementing the dbar algorithm for 2D difference EIT imaging
### 3.2 Package dependencies
| Packages | Optional | Note |
|----------------| ---------- |------------------------------------------|
| **numpy** | | tested with `numpy-1.19.1` |
| **scipy** | | tested with `scipy-1.5.0` |
| **matplotlib** | | tested with `matplotlib-3.3.2` |
| **trimesh** | | for loading external meshes |
| **pandas** | *Optional* | tested with `pandas-1.1.3` |
| **vispy** | *Optional* | failed with `vispy` in python 3.8 |
| **distmesh** | *Optional* | a build-in module is provided in `pyEIT` |
| **shapely** | *Optional* | for thorax mesh implementation |
**Q1, Why you choose vispy for 3D visualization?**
`pyEIT` uses `vispy` for visualizing 3D meshes (tetrahedron), and this module is not required if you are using 2D EIT only. `vispy` has minimal system dependencies and it is purely python. All you need is a decent graphical card with `OpenGL` support. It supports fast rendering, which I think is more superior to `vtk` or `mayavi` and it also has decent support for python 3. Please go to the website [vispy.org](http://vispy.org/) or github repository [vispy.github](https://github.com/vispy/vispy) for more details. Installation of vispy is simple by typing `python setup.py install`. We are also considering `mayavi` for a future version of `pyEIT`.
**Q2, When to use Shapely?**
`pyEIT` uses `Shapely` to build the thorax mesh and reconstruct EIT thoracic images. The thorax mesh shape is considered as a polygon and it is built based on real geometric measures given from a thorax figure simulated by `EIDORS` EIT MATLAB library. The figure in turns is based on a real thorax CT (Computed Tomography) scan.
To visualize thorax simulations, you should install `Shapely`. To do so, you should simply type `pip install Shapely` or `conda install shapely`. You can visit Shapely official documentation website [shapely.readthedocs.io](https://shapely.readthedocs.io/en/stable/index.html#) for more details or the dependency description at [Shapely.pypi](https://pypi.org/project/Shapely/).
## 4. Cite our work
`pyEIT` was published at the 17th International Conference on Electrical Impedance Tomography, 2016. It is now officially published at `softwareX`, vol (7), 2018.
**If you find `pyEIT` useful, please cite our work!**
```bibtex
@article{liu2018pyeit,
title={pyEIT: A python based framework for Electrical Impedance Tomography},
author={Liu, Benyuan and Yang, Bin and Xu, Canhua and Xia, Junying and Dai, Meng and Ji, Zhenyu and You, Fusheng and Dong, Xiuzhen and Shi, Xuetao and Fu, Feng},
journal={SoftwareX},
volume={7},
pages={304--308},
year={2018},
publisher={Elsevier}
}
```