https://github.com/lucasb-eyer/pydensecrf
Python wrapper to Philipp Krähenbühl's dense (fully connected) CRFs with gaussian edge potentials.
https://github.com/lucasb-eyer/pydensecrf
computer-vision crf cython eigen machine-learning pairwise-potentials unary-potentials
Last synced: 21 days ago
JSON representation
Python wrapper to Philipp Krähenbühl's dense (fully connected) CRFs with gaussian edge potentials.
- Host: GitHub
- URL: https://github.com/lucasb-eyer/pydensecrf
- Owner: lucasb-eyer
- License: mit
- Created: 2015-11-13T18:15:38.000Z (over 9 years ago)
- Default Branch: master
- Last Pushed: 2024-03-05T08:38:53.000Z (about 1 year ago)
- Last Synced: 2025-04-11T05:07:42.647Z (21 days ago)
- Topics: computer-vision, crf, cython, eigen, machine-learning, pairwise-potentials, unary-potentials
- Language: C++
- Size: 2.62 MB
- Stars: 1,973
- Watchers: 38
- Forks: 420
- Open Issues: 42
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
README
PyDenseCRF
==========
This is a (Cython-based) Python wrapper for [Philipp Krähenbühl's Fully-Connected CRFs](http://web.archive.org/web/20161023180357/http://www.philkr.net/home/densecrf) (version 2, [new, incomplete page](http://www.philkr.net/2011/12/01/nips/)).
If you use this code for your reasearch, please cite:
```
Efficient Inference in Fully Connected CRFs with Gaussian Edge Potentials
Philipp Krähenbühl and Vladlen Koltun
NIPS 2011
```
and provide a link to this repository as a footnote or a citation.
Installation
============
The package is on PyPI, so simply run `pip install pydensecrf` to install it.
If you want the newest and freshest version, you can install it by executing:
```
pip install git+https://github.com/lucasb-eyer/pydensecrf.git
```
and ignoring all the warnings coming from Eigen.
Note that you need a relatively recent version of Cython (at least version 0.22) for this wrapper,
the one shipped with Ubuntu 14.04 is too old. (Thanks to Scott Wehrwein for pointing this out.)
I suggest you use a [virtual environment](https://virtualenv.readthedocs.org/en/latest/) and install
the newest version of Cython there (`pip install cython`), but you may update the system version by
```
sudo apt-get remove cython
sudo pip install -U cython
```
### Problems on Windows/VS
Since this library needs to compile C++ code, installation can be a little more problematic than pure Python packages.
Make sure to [have Cython installed](https://github.com/lucasb-eyer/pydensecrf/issues/62#issuecomment-400563257) or try [installing via conda instead](https://github.com/lucasb-eyer/pydensecrf/issues/69#issuecomment-400639881) if you are getting problems.
PRs that improve Windows support are welcome.
### Problems on Colab/Kaggle Kernel
`pydensecrf` does not come pre-installed in Colab or Kaggle Kernel. Running `pip install pydensecrf` will result into
build failures. Follow these steps instead for Colab/Kaggle Kernel:
```
pip install -U cython
pip install git+https://github.com/lucasb-eyer/pydensecrf.git
```
Usage
=====
For images, the easiest way to use this library is using the `DenseCRF2D` class:
```python
import numpy as np
import pydensecrf.densecrf as dcrf
d = dcrf.DenseCRF2D(640, 480, 5) # width, height, nlabels
```
Unary potential
---------------
You can then set a fixed unary potential in the following way:
```python
U = np.array(...) # Get the unary in some way.
print(U.shape) # -> (5, 480, 640)
print(U.dtype) # -> dtype('float32')
U = U.reshape((5,-1)) # Needs to be flat.
d.setUnaryEnergy(U)
# Or alternatively: d.setUnary(ConstUnary(U))
```
Remember that `U` should be negative log-probabilities, so if you're using
probabilities `py`, don't forget to `U = -np.log(py)` them.
Requiring the `reshape` on the unary is an API wart that I'd like to fix, but
don't know how to without introducing an explicit dependency on numpy.
**Note** that the `nlabels` dimension is the first here before the reshape;
you may need to move it there before reshaping if that's not already the case,
like so:
```python
print(U.shape) # -> (480, 640, 5)
U = U.transpose(2, 0, 1).reshape((5,-1))
```
### Getting a Unary
There's two common ways of getting unary potentials:
1. From a hard labeling generated by a human or some other processing.
This case is covered by `from pydensecrf.utils import unary_from_labels`.
2. From a probability distribution computed by, e.g. the softmax output of a
deep network. For this, see `from pydensecrf.utils import unary_from_softmax`.
For usage of both of these, please refer to their docstrings or have a look at [the example](examples/inference.py).
Pairwise potentials
-------------------
The two-dimensional case has two utility methods for adding the most-common pairwise potentials:
```python
# This adds the color-independent term, features are the locations only.
d.addPairwiseGaussian(sxy=(3,3), compat=3, kernel=dcrf.DIAG_KERNEL, normalization=dcrf.NORMALIZE_SYMMETRIC)
# This adds the color-dependent term, i.e. features are (x,y,r,g,b).
# im is an image-array, e.g. im.dtype == np.uint8 and im.shape == (640,480,3)
d.addPairwiseBilateral(sxy=(80,80), srgb=(13,13,13), rgbim=im, compat=10, kernel=dcrf.DIAG_KERNEL, normalization=dcrf.NORMALIZE_SYMMETRIC)
```
Both of these methods have shortcuts and default-arguments such that the most
common use-case can be simplified to:
```python
d.addPairwiseGaussian(sxy=3, compat=3)
d.addPairwiseBilateral(sxy=80, srgb=13, rgbim=im, compat=10)
```
The parameters map to those in the paper as follows: `sxy` in the `Gaussian` case is `$\theta_{\gamma}$`,
and in the `Bilateral` case, `sxy` and `srgb` map to `$\theta_{\alpha}$` and `$\theta_{\beta}$`, respectively.
The names are shorthand for "x/y standard-deviation" and "rgb standard-deviation" and for reference, the formula is:
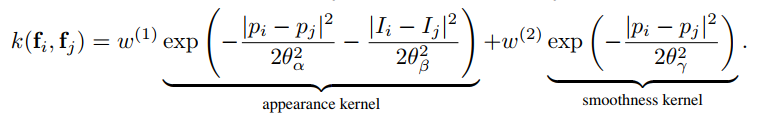
### Non-RGB bilateral
An important caveat is that `addPairwiseBilateral` only works for RGB images, i.e. three channels.
If your data is of different type than this simple but common case, you'll need to compute your
own pairwise energy using `utils.create_pairwise_bilateral`; see the [generic non-2D case](https://github.com/lucasb-eyer/pydensecrf#generic-non-2d) for details.
A good [example of working with Non-RGB data](https://github.com/lucasb-eyer/pydensecrf/blob/master/examples/Non%20RGB%20Example.ipynb) is provided as a notebook in the examples folder.
### Compatibilities
The `compat` argument can be any of the following:
- A number, then a `PottsCompatibility` is being used.
- A 1D array, then a `DiagonalCompatibility` is being used.
- A 2D array, then a `MatrixCompatibility` is being used.
These are label-compatibilites `µ(xi, xj)` whose parameters could possibly be [learned](https://github.com/lucasb-eyer/pydensecrf#learning).
For example, they could indicate that mistaking `bird` pixels for `sky` is not as bad as mistaking `cat` for `sky`.
The arrays should have `nlabels` or `(nlabels,nlabels)` as shape and a `float32` datatype.
### Kernels
Possible values for the `kernel` argument are:
- `CONST_KERNEL`
- `DIAG_KERNEL` (the default)
- `FULL_KERNEL`
This specifies the kernel's precision-matrix `Λ(m)`, which could possibly be learned.
These indicate correlations between feature types, the default implying no correlation.
Again, this could possiblty be [learned](https://github.com/lucasb-eyer/pydensecrf#learning).
### Normalizations
Possible values for the `normalization` argument are:
- `NO_NORMALIZATION`
- `NORMALIZE_BEFORE`
- `NORMALIZE_AFTER`
- `NORMALIZE_SYMMETRIC` (the default)
### Kernel weight
I have so far not found a way to set the kernel weights `w(m)`.
According to the paper, `w(2)` was set to 1 and `w(1)` was cross-validated, but never specified.
Looking through Philip's code (included in [pydensecrf/densecrf](https://github.com/lucasb-eyer/pydensecrf/tree/master/pydensecrf/densecrf)),
I couldn't find such explicit weights, and my guess is they are thus hard-coded to 1.
If anyone knows otherwise, please open an issue or, better yet, a pull-request.
Update: user @waldol1 has an idea in [this issue](https://github.com/lucasb-eyer/pydensecrf/issues/37). Feel free to try it out!
Inference
---------
The easiest way to do inference with 5 iterations is to simply call:
```python
Q = d.inference(5)
```
And the MAP prediction is then:
```python
map = np.argmax(Q, axis=0).reshape((640,480))
```
If you're interested in the class-probabilities `Q`, you'll notice `Q` is a
wrapped Eigen matrix. The Eigen wrappers of this project implement the buffer
interface and can be simply cast to numpy arrays like so:
```python
proba = np.array(Q)
```
Step-by-step inference
----------------------
If for some reason you want to run the inference loop manually, you can do so:
```python
Q, tmp1, tmp2 = d.startInference()
for i in range(5):
print("KL-divergence at {}: {}".format(i, d.klDivergence(Q)))
d.stepInference(Q, tmp1, tmp2)
```
Generic non-2D
--------------
The `DenseCRF` class can be used for generic (non-2D) dense CRFs.
Its usage is exactly the same as above, except that the 2D-specific pairwise
potentials `addPairwiseGaussian` and `addPairwiseBilateral` are missing.
Instead, you need to use the generic `addPairwiseEnergy` method like this:
```python
d = dcrf.DenseCRF(100, 5) # npoints, nlabels
feats = np.array(...) # Get the pairwise features from somewhere.
print(feats.shape) # -> (7, 100) = (feature dimensionality, npoints)
print(feats.dtype) # -> dtype('float32')
dcrf.addPairwiseEnergy(feats)
```
In addition, you can pass `compatibility`, `kernel` and `normalization`
arguments just like in the 2D gaussian and bilateral cases.
The potential will be computed as `w*exp(-0.5 * |f_i - f_j|^2)`.
### Pairwise potentials for N-D
User @markusnagel has written a couple numpy-functions generalizing the two
classic 2-D image pairwise potentials (gaussian and bilateral) to an arbitrary
number of dimensions: `create_pairwise_gaussian` and `create_pairwise_bilateral`.
You can access them as `from pydensecrf.utils import create_pairwise_gaussian`
and then have a look at their docstring to see how to use them.
Learning
--------
The learning has not been fully wrapped. If you need it, get in touch or better
yet, wrap it and submit a pull-request!
Here's a pointer for starters: issue#24. We need to wrap the gradients and getting/setting parameters.
But then, we also need to do something with these, most likely call [minimizeLBFGS from optimization.cpp](https://github.com/lucasb-eyer/pydensecrf/blob/d824b89ee3867bca3e90b9f04c448f1b41821524/pydensecrf/densecrf/src/optimization.cpp).
It should be relatively straightforward to just follow the learning examples included in the [original code](http://graphics.stanford.edu/projects/drf/densecrf_v_2_2.zip).
Common Problems
===============
`undefined symbol` when importing
---------------------------------
If while importing pydensecrf you get an error about some undefined symbols (for example `.../pydensecrf/densecrf.so: undefined symbol: _ZTINSt8ios_base7failureB5cxx11E`), you most likely are inadvertently mixing different compilers or toolchains. Try to see what's going on using tools like `ldd`. If you're using Anaconda, [running `conda install libgcc` might be a solution](https://github.com/lucasb-eyer/pydensecrf/issues/28).
ValueError: Buffer dtype mismatch, expected 'float' but got 'double'
--------------------------------------------------------------------
This is a pretty [co](https://github.com/lucasb-eyer/pydensecrf/issues/52)mm[on](https://github.com/lucasb-eyer/pydensecrf/issues/49) user error.
It means exactly what it says: you are passing a `double` but it wants a `float`.
Solve it by, for example, calling `d.setUnaryEnergy(U.astype(np.float32))` instead of just `d.setUnaryEnergy(U)`, or using `float32` in your code in the first place.
My results are all pixelated like [MS Paint's airbrush tool](http://lmgtfy.com/?q=MS+Paint+Airbrush+tool)!
----------------------------------------------------------
You screwed up reshaping somewhere and treated the class/label dimension as spatial dimension.
This is you misunderstanding NumPy's memory layout and nothing that PyDenseCRF can detect or help with.
This mistake often happens for the Unary, see the [**Note** in that section of the README](https://github.com/lucasb-eyer/pydensecrf#unary-potential).
Maintaining
===========
These are instructions for maintainers about how to release new versions. (Mainly instructions for myself.)
```
# Go increment the version in setup.py
> python setup.py build_ext
> python setup.py sdist
> twine upload dist/pydensecrf-VERSION_NUM.tar.gz
```
And that's it. At some point, it would be cool to automate this on [TravisCI](https://docs.travis-ci.com/user/deployment/pypi/), but not worth it yet.
At that point, looking into [creating "manylinux" wheels](https://github.com/pypa/python-manylinux-demo) might be nice, too.
Testing
=======
Thanks to @MarvinTeichmann we now have proper tests, install the package and run `py.test`.
Maybe there's a better way to run them, but both of us don't know :smile: