Ecosyste.ms: Awesome
An open API service indexing awesome lists of open source software.
https://github.com/m-pilia/disptools
Generate displacement fields with known volume changes
https://github.com/m-pilia/disptools
cuda image-processing jacobian python3
Last synced: 2 months ago
JSON representation
Generate displacement fields with known volume changes
- Host: GitHub
- URL: https://github.com/m-pilia/disptools
- Owner: m-pilia
- License: mit
- Created: 2018-03-19T09:44:06.000Z (almost 7 years ago)
- Default Branch: master
- Last Pushed: 2024-09-06T18:04:22.000Z (5 months ago)
- Last Synced: 2024-11-07T12:06:29.808Z (3 months ago)
- Topics: cuda, image-processing, jacobian, python3
- Language: C
- Homepage: https://martinopilia.com/disptools
- Size: 7.29 MB
- Stars: 23
- Watchers: 4
- Forks: 3
- Open Issues: 7
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
README
disptools
=========
Generate displacement fields with known volume changes
------------------------------------------------------
[](https://github.com/m-pilia/disptools/releases/latest)
[](https://pypi.python.org/pypi/disptools)
[](https://pypi.org/project/disptools)
[](https://github.com/m-pilia/disptools/blob/master/LICENSE)
[](https://github.com/m-pilia/disptools/actions/workflows/checks_linux.yml)
[](https://codecov.io/gh/m-pilia/disptools/branch/master)
[](https://pepy.tech/project/disptools)
This library provides utilities to generate and manipulate displacement fields with known volume changes. It implements three search-based algorithms for the generation of deformation fields, along with a small collection of utility functions, and provides optional GPU acceleration through a CUDA implementation.
The three algorithms implemented are referred as:
+ gradient: a gradient descent method (default).
+ greedy: a greedy search method proposed in [[1]](#1).
+ matching: a volume matching method proposed in [[2]](#2) and [[3]](#3). The implementation comes from the [PREDICT atrophysim tool](https://www.nitrc.org/projects/atrophysim).
The library is built on top of SimpleITK, in order to provide a simple yet powerful set of functionalities for image analysis. Images stored as numpy arrays can be easily converted from and to [SimpleITK](http://simpleitk.github.io/SimpleITK-Notebooks/01_Image_Basics.html) and [ITK](https://blog.kitware.com/convert-itk-data-structures-to-numpy-arrays/) image objects.
### Documentation
The complete documentation for this package is available on https://martinopilia.com/disptools.
### Quick example
```python
import SimpleITK as sitk
import disptools.displacements as dsp
import disptools.drawing as drw
# Create an example Jacobian map
# A spherical ROI with a Jacobian of 1.1 (expansion)
jacobian = drw.create_sphere(10, 40, fg_val=1.1, bg_val=1.0)
# Create a binary mask for the ROI
mask = drw.create_sphere(10, 40) > 0
# Generate the displacement
displacement = dsp.displacement(jacobian, mask=mask)
# Check the correctness of the result within the ROI
error = jacobian - dsp.jacobian(displacement)
error = sitk.Mask(error, mask)
```
A 3D rendering of the resulting displacement field with [ParaView](https://www.paraview.org/), and a visualisation of the the error on the Jacobian within the ROI:
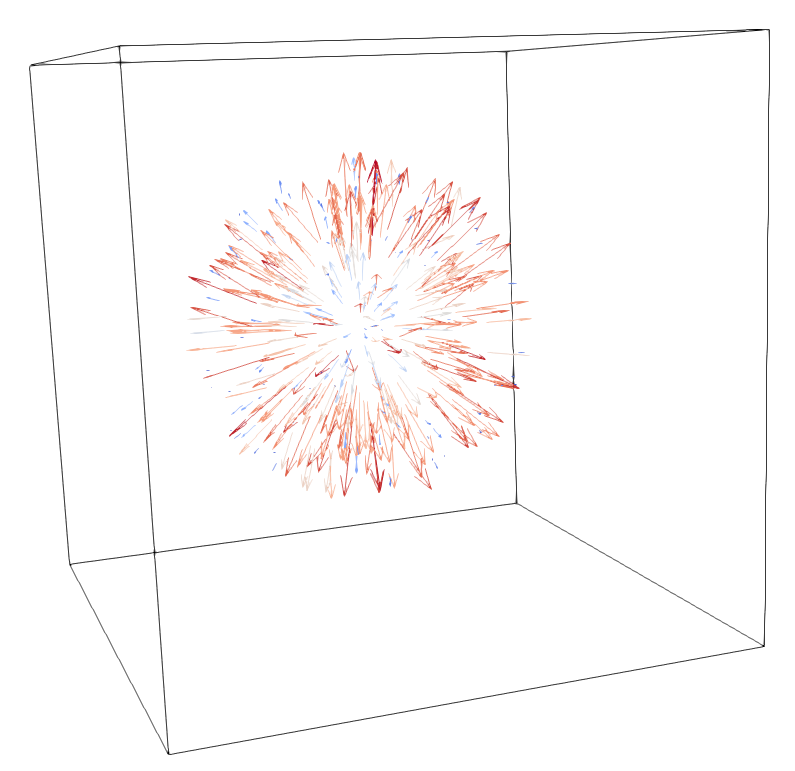
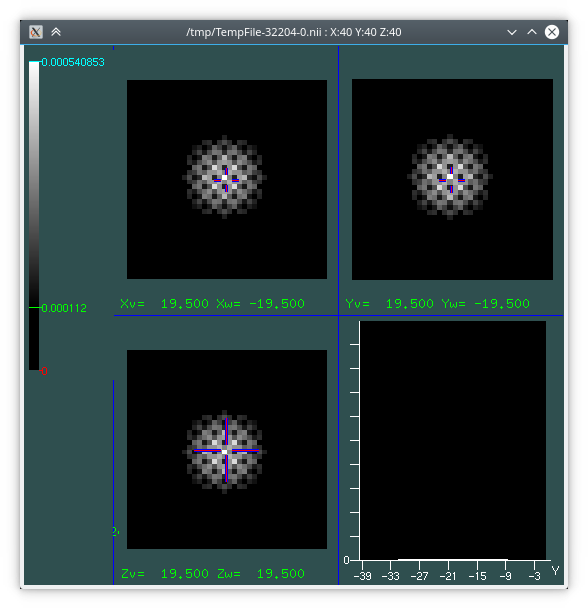
### Architecture
The project is structured in three layers:
+ a pure standard [C99](https://en.wikipedia.org/wiki/C99) library (whose headers are in src/headers), with no external dependencies, implementing the core algorithms for the generation of deformation fields. It is a standalone library that can be directly included in a C or C++ project. It is paired with an optional [CUDA](https://en.wikipedia.org/wiki/CUDA) library, whose headers are in cuda/headers, that depends on src/headers and provides a GPGPU implementation of the key routines.
+ a [Python C extension](https://docs.python.org/3.6/extending/extending.html) package _disptools (whose source is in the file python_c_extension/_disptools.c), providing a bare Python wrapper to the aforementioned library, using the [NumPy C API](https://docs.scipy.org/doc/numpy-1.14.0/reference/c-api.html) to pass arrays. This can be directly included in a Python project with no dependencies other than NumPy.
+ a Python package (disptools), that wraps the _disptools package providing file IO (through SimpleITK) and implementing high-level features (such as the multi-resolution framework) and auxiliary utilities and functions.
- disptools.displacements: module providing the main functions for the generation and manipulation of displacement fields.
- disptools.drawing: collection of utilities to create test images.
- disptools.io: collection of utilities to read and write to file.
- disptools.measure: collection of utilities to measure some image features.
- disptools.simulatrophy: routines to interface with the [Simul@atrophy](https://github.com/Inria-Asclepios/simul-atrophy) tool.
- disptools.predict: routines to interface with the [PREDICT](https://www.nitrc.org/projects/atrophysim) tool.
### Install
This package is available on [PyPI](https://pypi.org/project/disptools) both as source distribution and as a Windows pre-compiled binary wheel. You can install it with pip:
```bash
python3 -m pip install disptools
```
As always, it is recommended to use the package inside a [virtual environment](https://docs.python.org/3/tutorial/venv.html).
### Build from source
#### Requirements
Requirements are specified by the requirements.txt file and can be installed with pip.
```bash
python3 -m pip install -r requirements.txt
```
The library is a cross-platform Python 3.5+ package, with a compiled C extension. The Python dependencies are:
+ [numpy](https://github.com/numpy/numpy) ([pypi package](https://pypi.python.org/pypi/numpy))
+ [scipy](https://github.com/scipy/scipy) ([pypi package](https://pypi.org/pypi/scipy))
+ [SimpleITK](https://github.com/SimpleITK/SimpleITK) ([pypi package](https://pypi.org/pypi/SimpleITK))
Build dependencies are a standard C compiler (tested with gcc 8.2 on Linux, mingw-w64 7.2 and MSVC 19 on Windows 10), [CMake](https://cmake.org/), the [numpy](https://pypi.python.org/pypi/numpy) and the [setuptools](https://pypi.python.org/pypi/setuptools) packages. [scikit-build](https://pypi.python.org/pypi/scikit-build) may be required to build the other Python dependencies.
Some optional dependencies are required only for a limited set of features, and the package should build and run without them:
+ [itk](https://github.com/InsightSoftwareConsortium/ITK) ([pypi package](https://pypi.org/project/itk)): for disptools.drawing.sitk_to_itk
+ [vtk](https://github.com/Kitware/VTK) ([pypi package](https://pypi.org/project/vtk)): for disptools.io.write_vtk_points
+ [ply](https://github.com/dabeaz/ply) ([pypi package](https://pypi.org/project/ply)): for disptools.io.read_elastix_parameters
+ [scikit-image](https://github.com/scikit-image/scikit-image) ([pypi package](https://pypi.org/project/scikit-image)): for some features of disptools.drawing.extract_slice, and to run the unit tests
#### Build options
The following environment variables affect the setup.py:
+ DISPTOOLS_OPT=ON: enable non-portable optimisations.
+ DISPTOOLS_DEBUG=ON: disable optimisations, compile with debug symbols.
+ DISPTOOLS_CUDA_SUPPORT=ON: enable CUDA support.
#### Windows (Visual Studio) and Linux
Install the dependencies with your favourite package manager. For example, with pip:
```bash
python3 -m pip install -r requirements.txt
```
The package provides a setuptools based install script. To install the library, run from the project root folder
```bash
python3 setup.py install
```
which should build the C extension and install the Python package.
#### Windows (MinGW)
1. First, be sure that [mingw](https://mingw-w64.org), CMake and Python are installed and their executables [are in your PATH](https://docs.python.org/3/using/windows.html#excursus-setting-environment-variables).
2. Ensure that gcc is working correctly:
```none
> gcc --version
gcc (x86_64-posix-seh-rev1, Built by MinGW-W64 project) 7.2.0
Copyright (C) 2017 Free Software Foundation, Inc.
This is free software; see the source for copying conditions. There is NO
warranty; not even for MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE.
```
3. Ensure that distutils correctly recognises your version of Visual Studio (even when using mingw). Open the file C:\Users\yourname\AppData\Local\Programs\Python\Python3x\Lib\distutils\cygwinccompiler.py (the exact location may vary according to your setup) and check that your version of Visual Studio is present in the function get_msvcr(); if not, adjust it according to the following:
```python
def get_msvcr():
"""Include the appropriate MSVC runtime library if Python was built
with MSVC 7.0 or later.
"""
msc_pos = sys.version.find('MSC v.')
if msc_pos != -1:
msc_ver = sys.version[msc_pos+6:msc_pos+10]
if msc_ver == '1300':
# MSVC 7.0
return ['msvcr70']
elif msc_ver == '1310':
# MSVC 7.1
return ['msvcr71']
elif msc_ver == '1400':
# VS2005 / MSVC 8.0
return ['msvcr80']
elif msc_ver == '1500':
# VS2008 / MSVC 9.0
return ['msvcr90']
elif msc_ver == '1600':
# VS2010 / MSVC 10.0
return ['msvcr100']
elif msc_ver == '1700':
# Visual Studio 2012 / Visual C++ 11.0
return ['msvcr110']
elif msc_ver == '1800':
# Visual Studio 2013 / Visual C++ 12.0
return ['msvcr120']
elif msc_ver == '1900':
# Visual Studio 2015 / Visual C++ 14.0
# "msvcr140.dll no longer exists" http://blogs.msdn.com/b/vcblog/archive/2014/06/03/visual-studio-14-ctp.aspx
return ['vcruntime140']
else:
raise ValueError("Unknown MS Compiler version %s " % msc_ver)
```
4. Ensure that the library vcruntime140.dll is present in your library path. Otherwise, download it and place it in C:\Users\yourname\AppData\Local\Programs\Python\Python3x\libs (the exact path may vary according to your setup).
5. Clone the sources of this package with git, or download and extract them as a zip archive. Move to the root folder of the sources (C:\Users\yourname\disptools in this example), specify the right compiler, and launch the setup script to build and install the package.
```cmd
> cd C:\Users\yourname\disptools
> python setup.py setopt --command=build --option=compiler --set-value=mingw32
> python -m pip install -r requirements.txt
> python setup.py install
```
### References
+ [1] van Eede, M. C., Scholz, J., Chakravarty, M. M., Henkelman, R. M., and Lerch, J. P. *Mapping registration sensitivity in MR mouse brain images.* Neuroimage 82 (2013), 226–236.
+ [2] Karaçali, B., and Davatzikos, C. *Estimating topology preserving and smooth displacement fields.* IEEE Transactions on Medical Imaging 23, 7 (2004), 868–880.
+ [3] Karaçali, B., and Davatzikos, C. *Simulation of tissue atrophy using a topology preserving transformation model.* IEEE transactions on medical imaging 25, 5 (2006), 649–652.
### License
The software is distributed under the MIT license.