Ecosyste.ms: Awesome
An open API service indexing awesome lists of open source software.
https://github.com/maastrichtu-ids/dsri-workshop-start-app
⚡ Short workshop to start Data Science applications on the Data Science Research Infrastructure at Maastricht University
https://github.com/maastrichtu-ids/dsri-workshop-start-app
data-science data-science-research dsri openshift workshop
Last synced: 13 days ago
JSON representation
⚡ Short workshop to start Data Science applications on the Data Science Research Infrastructure at Maastricht University
- Host: GitHub
- URL: https://github.com/maastrichtu-ids/dsri-workshop-start-app
- Owner: MaastrichtU-IDS
- License: cc0-1.0
- Created: 2020-12-01T19:18:38.000Z (about 4 years ago)
- Default Branch: main
- Last Pushed: 2021-11-22T17:44:27.000Z (about 3 years ago)
- Last Synced: 2024-04-18T06:14:50.359Z (9 months ago)
- Topics: data-science, data-science-research, dsri, openshift, workshop
- Homepage: https://maastrichtu-ids.github.io/dsri-workshop-start-app
- Size: 66.4 KB
- Stars: 4
- Watchers: 6
- Forks: 1
- Open Issues: 0
-
Metadata Files:
- Readme: README.md
- Contributing: CONTRIBUTING.md
- License: LICENSE
Awesome Lists containing this project
README
A workshop to get started with the [**Data Science Research Infrastructure (DSRI)**](https://maastrichtu-ids.github.io/dsri-documentation/) in an hour 🕐 (hopefully)!
During this workshop, you will:
* Access the Data Science Research Infrastructure web UI
* Create a new application from a template in the catalog (RStudio, JupyterLab or VScode )
* Access the application
* Add source code and data in the application
* Optionally install the `oc` command line interface
Prerequisites:
* A web browser (Chrome preferably, as some other web browsers have issues with the VSCode terminal)
* An account on the DSRI with your UM email
* Access to the [**UM VPN**](https://vpn.maastrichtuniversity.nl), or direct connection to **UMnet** or **eduroam** at Maastricht University
* Students can use the Athena Student Desktop at [athenadesktop.maastrichtuniversity.nl](https://athenadesktop.maastrichtuniversity.nl) to access the DSRI web UI
---
## Access the DSRI 🔑
📖 The DSRI documentation can be found at [https://maastrichtu-ids.github.io/dsri-documentation](https://maastrichtu-ids.github.io/dsri-documentation)
1. Connect to the [**UM VPN**](https://vpn.maastrichtuniversity.nl).
- Students can use the Athena Student Desktop at [athenadesktop.maastrichtuniversity.nl](https://athenadesktop.maastrichtuniversity.nl) to access the DSRI web UI
- On Linux you can use [`openconnect`](https://websiteforstudents.com/install-openconnect-ssl-vpn-client-on-ubuntu-18-04-18-04/):
```bash
sudo openconnect --passwd-on-stdin -u YOUR.UM.USER --authgroup 01-Employees vpn-rw1.maastrichtuniversity.nl
```
2. Access the DSRI [OpenShift](https://www.okd.io/) web UI
- 📖 See the [complete documentation to access the DSRI](https://maastrichtu-ids.github.io/dsri-documentation/docs/access-dsri)
3. 👩💻 Go to the [**workspace-workshop** project](https://console-openshift-console.apps.dsri2.unimaas.nl/topology/ns/workspace-workshop/graph) in the OpenShift web UI
---
## Start an application 🚀
Start a **JupyterLab/RStudio/VSCode** application from the DSRI catalog in ids-projects
📖 See how to deploy [JupyterLab](https://maastrichtu-ids.github.io/dsri-documentation/docs/deploy-jupyter), [RStudio](https://maastrichtu-ids.github.io/dsri-documentation/docs/deploy-rstudio), [VSCode](https://maastrichtu-ids.github.io/dsri-documentation/docs/deploy-vscode) and lots more.
1. 👨💻 Use your name to generate a unique **Application name**, e.g. `rstudio-vemonet`
2. Persistent storage will create automatically.
- It can be found at [https://console-openshift-console.apps.dsri2.unimaas.nl/k8s/cluster/persistentvolumes](https://console-openshift-console.apps.dsri2.unimaas.nl/k8s/cluster/persistentvolumes)
- ⚠️ When copy/pasting the storage name it can happen that a space is added at the end. **Be careful to trim all spaces at the start and the end of the storage name** before starting the application, otherwise it will fail
3. Access the application you just started
- 👩💻 You can find the URL of your application in the [OpenShift web UI **workshops** overview](https://console-openshift-console.apps.dsri2.unimaas.nl/topology/ns/workspace-workshop).
---
## Upload files 🗂️
👨💻 For small and medium size files you can simply drag and drop files and folder in the application web UI, or use the **Upload files** button in RStudio.
> This solution works for files up to a few hundred MBs (depending on the application, use it until it fails!).
---
## Upload your code 📜
We recommend you to use [`git`](https://git-scm.com/) with GitHub or GitLab, you can use it directly from the terminal in all applications, or use the web UI integration each app proposes.
📖 See the documentation for each application:
* RStudio: [https://maastrichtu-ids.github.io/dsri-documentation/docs/deploy-rstudio#use-git-in-rstudio](https://maastrichtu-ids.github.io/dsri-documentation/docs/deploy-rstudio#use-git-in-rstudio)
* VSCode: [https://maastrichtu-ids.github.io/dsri-documentation/docs/deploy-vscode#use-git-in-vscode](https://maastrichtu-ids.github.io/dsri-documentation/docs/deploy-vscode#use-git-in-vscode)
* JupyterLab (with `jupyterlab-git` extension installed): [https://maastrichtu-ids.github.io/dsri-documentation/docs/deploy-jupyter#use-git-in-jupyterlab](https://maastrichtu-ids.github.io/dsri-documentation/docs/deploy-jupyter#use-git-in-jupyterlab)
---
## Upload large data files 📦
For large data files you will need to [install the `oc` command line interface](https://maastrichtu-ids.github.io/dsri-documentation/docs/openshift-install).
> If you have the time it can be quickly installed on MacOS, Linux (works with WSL):
>
> * On **Linux** 🐧
>
> ```bash
> wget https://github.com/openshift/origin/releases/download/v3.11.0/openshift-origin-client-tools-v3.11.0-0cbc58b-linux-64bit.tar.gz
> tar xvf openshift-origin-client-tools*.tar.gz
> cd openshift-origin-client*/
> sudo mv oc kubectl /usr/local/bin/
> ```
>
> * On **Mac** 🍎
>
> ```bash
> brew install openshift-cli
> ```
>
> * On **Windows** 🏢
> * [Checkout the installation documentation](https://maastrichtu-ids.github.io/dsri-documentation/docs/openshift-install#on-windows).
📖 See the [complete documentation to upload large data file](https://maastrichtu-ids.github.io/dsri-documentation/docs/openshift-load-data)
> 💡 You will have a better connection when directly connected to the UMnet network (or eduroam at UM) to upload large data file. Even better if you can use ethernet wires.
---
## Stop and delete your application ❌
👨💻 Stop your application from the [OpenShift web UI **Topology** page](https://console-openshift-console.apps.dsri2.unimaas.nl/topology/ns/workspace-workshop/graph):
> You can use the *Filter by name* search box to quickly find your application based on the name you gave it.
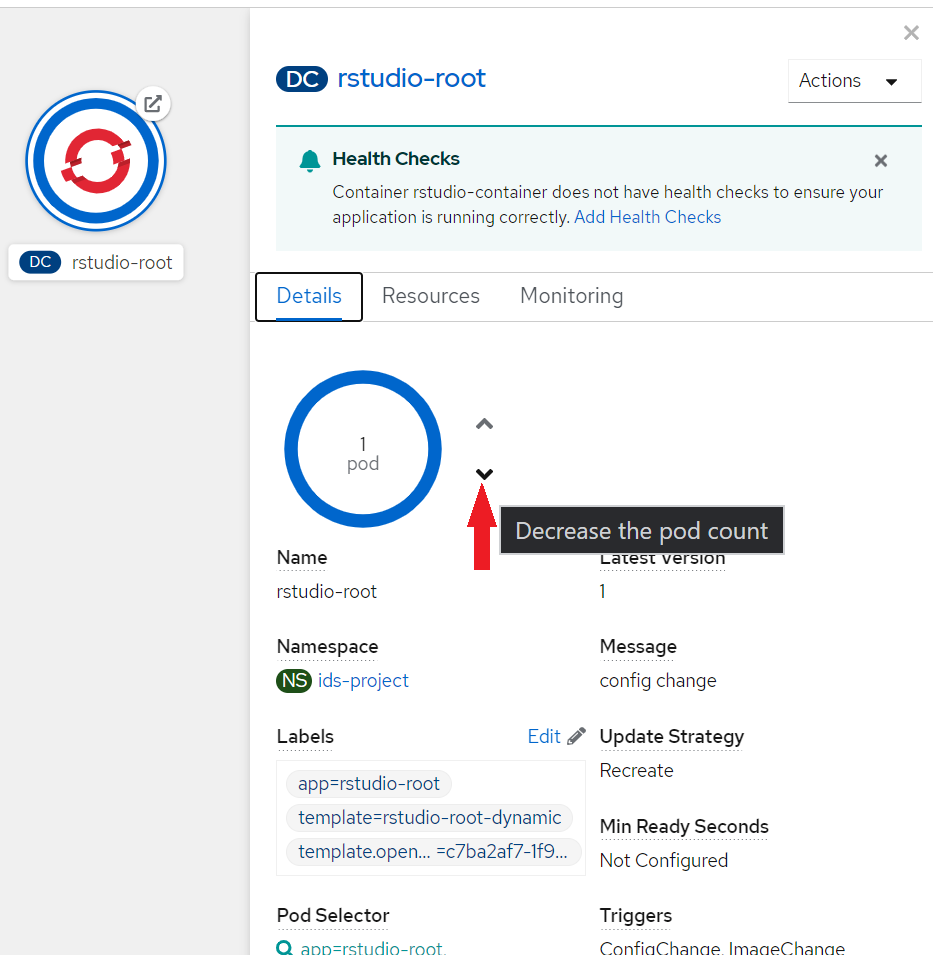
> Note: creating more than one pod ("Scale up") is useless for most data science applications, such as RStudio, VSCode or JupyterLab. It is only relevant for applications running as a cluster, like Apache Flink or Apache Spark, or web application with a lot of traffic (OpenShift will redirect the traffic depending on pod availability, and start new pods if required, aka. horizontal scaling).
👩💻 Delete your application:
* If you installed the `oc` command line interface, it is easier to use it to delete all the objects related to your application:
```bash
oc delete all,secret,configmaps,serviceaccount,rolebinding --selector app=my-application
```
> Replace `my-application` by the Application name you defined.
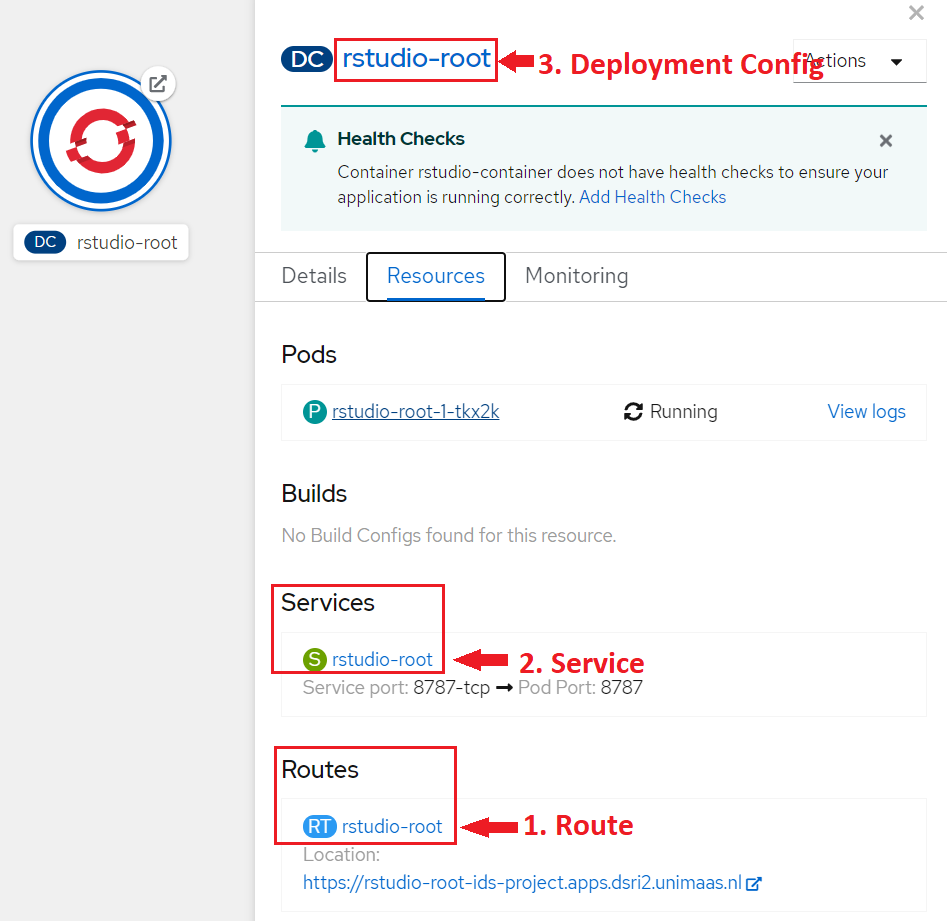
* Otherwise you will need to manually delete a few objects related to your application in the OpenShift web UI, it can be done easily from the **Overview** page:
1. Delete the **Route**
2. Delete the **Service**
3. Delete the **Deployment Config**
📖 See the [complete documentation to delete an application](https://maastrichtu-ids.github.io/dsri-documentation/docs/openshift-delete-services#delete-an-application).
---
## See you soon! 👋
[📝 Fill this form](https://docs.google.com/forms/d/e/1FAIpQLSdndn0naNmj2ACpLE5j1S3Ngb1PCXK_Gl7oB-hI_mN4Z_NBQw/viewform) to help us create a project for you on the Data Science Research Infrastructure for a longer term!