https://github.com/manmolecular/http-response-clustering
:chart_with_downwards_trend: Clustering of HTTP responses using k-means++ and the elbow method
https://github.com/manmolecular/http-response-clustering
data-analysis elbow-method elbow-plot jupyter k-means-plus-plus python3
Last synced: 5 months ago
JSON representation
:chart_with_downwards_trend: Clustering of HTTP responses using k-means++ and the elbow method
- Host: GitHub
- URL: https://github.com/manmolecular/http-response-clustering
- Owner: manmolecular
- License: gpl-2.0
- Created: 2020-03-22T07:41:09.000Z (over 5 years ago)
- Default Branch: master
- Last Pushed: 2021-07-21T18:14:23.000Z (over 4 years ago)
- Last Synced: 2025-03-06T05:44:33.934Z (9 months ago)
- Topics: data-analysis, elbow-method, elbow-plot, jupyter, k-means-plus-plus, python3
- Language: Jupyter Notebook
- Homepage:
- Size: 959 KB
- Stars: 2
- Watchers: 2
- Forks: 0
- Open Issues: 2
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
README
# http-response-clustering
[](https://en.wikipedia.org/wiki/Linux)
[](https://www.python.org/downloads/)
[](https://github.com/manmolecular/http-response-clustering/issues)
[](https://github.com/manmolecular/http-response-clustering/pulls)
[](https://github.com/manmolecular/http-response-clustering/commits/master)
:chart_with_downwards_trend: Clustering of HTTP responses using k-means++ and the elbow method (PoC)
## Screenshot
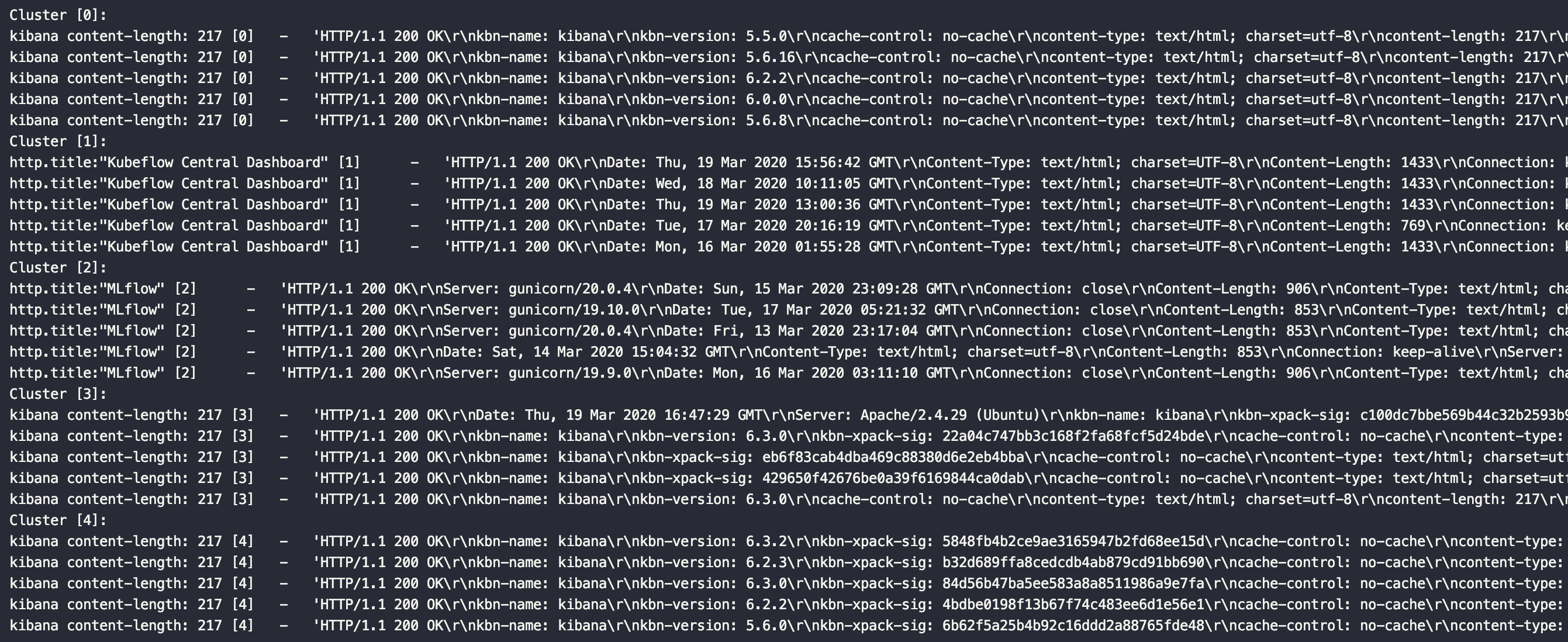
A basic example of dividing HTTP responses into clusters
## Contents
1. [Disclaimer](#disclaimer)
1. [Description](#description)
1. [Examples](#examples)
1. [Requirements](#requirements)
1. [Installation](#installation)
1. [Determine the number of clusters](#determine-the-number-of-clusters)
1. [Provide your data into clusters](#provide-your-data-into-clusters)
1. [Format of the HTTP responses to analyze](#format-of-the-http-responses-to-analyze)
## Disclaimer
:construction: This work is just an early-proof-of-concept, and almost in deep WIP status. So feel free to make some issues, feature requests, and bug reports. Thanks.
## Description
This module based on SciPy (SciKit) implementation of K-means (to be more precious, K-means++) clustering method allows you to sort different HTTP responses from various HTTP hosts into some clusters, divided by several unique features.
This can be useful when, for example, you cannot process a large amount of data from various network scanners (for example, Nmap Network Scanner, Shodan database, Censys database, etc.) or collected by yourself, because the quantity is too big and responses look almost the same.
In this case, you can try to divide all of your data into some small subgroups called clusters to find some unique features in every group.
## Examples
See [interactive_cluster.ipynb](/interactive_cluster.ipynb) notebook
## Requirements
### Run in docker
- Docker
- docker-compose
### Run on host directly
- Python 3.7+
- pip3
## Installation
### Run in docker
```
docker-compose up
```
The Jupyter notebook will be running on the `http://localhost:8888/`, follow the instructions from your terminal logs
**Note:** this repository contains 2 Dockerfiles: one is based on the Debian default python image and another one based on the Alpine python image. You can choose a preferable variant via modifying `docker-compose.yml` file:
replace `docker/debian/Dockerfile` string in `docker-compose.yml` file with `docker/alpine/Dockerfile`, if you want to build the image on the Alpine base.
By default, the Debian python image will be built.
### Run on host
Install the requirements and you are ready to go:
```
pip3 install -r requirements.txt
```
After that, you can use the Jupyter Notebook or module itself directly with `interactive_cluster.ipynb` notebook:
```
jupyter notebook
```
or with `clustering.py`:
```
python3 clustering.py
```
## Determine the number of clusters
One of the problems here is to understand how many clusters can we get from our results? One, three, five, or maybe even ten different clusters? In case when we know the quantity (for example, when we have 1.000 hosts - half of them is Apache servers, and another half - is Nginx servers, and we only need to divide these hosts into 2 clusters), it can be pretty easy - we just need to define the number of clusters as 2, for example. But in the case when we don't know exactly the quantity, we need to use some additional method to understand possible effective numbers of clusters. To do this we use "The Elbow Method" - heuristic method of data validation and analysis that helps to find the right quantity of clusters.
To demonstrate this method, you can play around with `interactive_cluster.ipynb` notebook.
## Provide your data into clusters
To try these methods with your data, you can add collected HTTP-responses in the format that you can find in the "data" directory.
In two words, the format must be the following:
```json
{
"my-class-1":[
"response-1",
"response-2",
"response-3"
],
"my-class-2":[
"response-1",
"response-2"
]
}
```
`"my-class-1"` key can be anything you want, if you don't have any predictions:
```json
{
"any":[
"response-1",
"response-2",
"response-3",
"response-4"
]
}
```
You need to create the JSON file with your data, provide a name for it (for example, `"hosts_example.json"`), put your data in the format above these lines and provide the name of your file into files handler:
```python3
raw_results = FilesHandler().open_results(filename="data/hosts_example.json")
```
## Format of the HTTP responses to analyze
Format of the HTTP responses must the like raw string with all the special characters, like, for example, from cURL:
```
HTTP/1.1 200 OK\r\nkbn-name: kibana\r\nkbn-version: 6.3.2\r\nkbn-xpack-sig: 242104651e8721ad01d1a77b0e87738e\r\ncache-control: no-cache\r\ncontent-type: text/html; charset=utf-8\r\ncontent-length: 217\r\naccept-ranges: bytes\r\nvary: accept-encoding\r\nDate: Thu, 19 Mar 2020 15:30:33 GMT\r\nConnection: keep-alive\r\n\r\n
```