https://github.com/mlin/sqlite_zstd_vfs
SQLite3 extension for read/write storage compression with Zstandard
https://github.com/mlin/sqlite_zstd_vfs
compression-formats sqlite sqlite3 zstandard zstd
Last synced: 10 months ago
JSON representation
SQLite3 extension for read/write storage compression with Zstandard
- Host: GitHub
- URL: https://github.com/mlin/sqlite_zstd_vfs
- Owner: mlin
- License: apache-2.0
- Created: 2020-05-24T08:20:57.000Z (over 5 years ago)
- Default Branch: main
- Last Pushed: 2024-01-13T22:48:31.000Z (about 2 years ago)
- Last Synced: 2025-03-31T10:01:34.301Z (10 months ago)
- Topics: compression-formats, sqlite, sqlite3, zstandard, zstd
- Language: C++
- Size: 27.4 MB
- Stars: 173
- Watchers: 9
- Forks: 10
- Open Issues: 1
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
README
# sqlite_zstd_vfs
This [SQLite VFS extension](https://www.sqlite.org/vfs.html) provides streaming storage compression with [Zstandard](https://facebook.github.io/zstd/), transparently compressing [pages of the main database file](https://www.sqlite.org/fileformat.html) as they're written out, and later decompressing them as they're read in. It runs page de/compression on background threads and occasionally generates [dictionaries](https://github.com/facebook/zstd#the-case-for-small-data-compression) to improve subsequent compression.
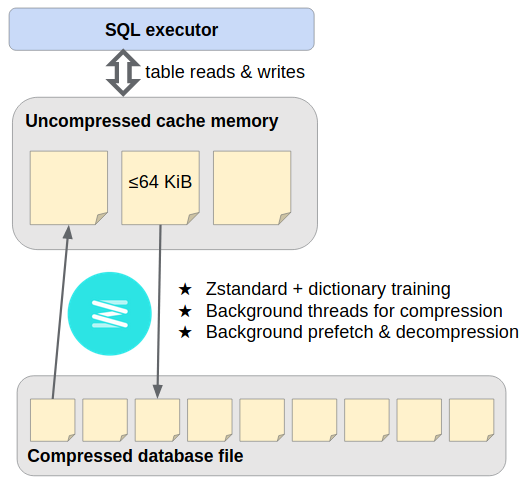
Compressed page storage operates similarly to the [design of ZIPVFS](https://sqlite.org/zipvfs/doc/trunk/www/howitworks.wiki), the SQLite developers' proprietary extension. Because we're not as familiar with the internal "pager" module, we use a full-fledged SQLite database as the bottom-most layer. Where SQLite would write database page #P at offset P × page_size in the disk file, instead we `INSERT INTO outer_page_table(rowid,data) VALUES(P,compressed_inner_page)`, and later `SELECT data FROM outer_page_table WHERE rowid=P`. *You mustn't be afraid to dream a little smaller, darling...*
**USE AT YOUR OWN RISK:** The extension makes fundamental changes to the database storage layer. While designed to preserve ACID transaction safety, it's young and unlikely to have zero bugs. This project is not associated with the SQLite developers.
### Included in GenomicSQLite
**The easiest way to use the VFS is through the [Genomics Extension for SQLite](https://github.com/mlin/GenomicSQLite), which bundles it as a core component with pre-tuned settings and nicely-packaged language bindings. That extension's bioinformatics-specific features can be ignored without harm.**
Standalone instructions follow.
## Quick start
Prerequisites:
* C++11 compiler, CMake >= 3.11
* *Up-to-date* packages for SQLite3, Zstandard, and libcurl development, e.g. Ubuntu 20.04 `sqlite3 libsqlite3-dev libzstd-dev libcurl4-openssl-dev`
Fetch source code and compile the [SQLite loadable extension](https://www.sqlite.org/loadext.html):
```
git clone https://github.com/mlin/sqlite_zstd_vfs.git
cd sqlite_zstd_vfs
cmake -DCMAKE_BUILD_TYPE=Release -B build
cmake --build build -j $(nproc)
```
Download a ~1MB example database and use the `sqlite3` CLI to create a compressed version of it:
```
wget https://github.com/lerocha/chinook-database/raw/master/ChinookDatabase/DataSources/Chinook_Sqlite.sqlite
sqlite3 Chinook_Sqlite.sqlite -bail \
-cmd '.load build/zstd_vfs.so' \
"VACUUM INTO 'file:Chinook_Sqlite.zstd.sqlite?vfs=zstd'"
ls -l Chinook_Sqlite.*
```
The compressed version is about 40% smaller; you'll often see better, with larger databases and some tuning (see below). You can start a new database with `sqlite3 -cmd '.load build/zstd_vfs.so' -cmd '.open file:new_db?vfs=zstd'` but the above approach provides this example something to work with.
Query the compressed database:
```
sqlite3 :memory: -bail \
-cmd '.load build/zstd_vfs.so' \
-cmd '.open file:Chinook_Sqlite.zstd.sqlite?mode=ro&vfs=zstd' \
"select e.*, count(i.invoiceid) as 'Total Number of Sales'
from employee as e
join customer as c on e.employeeid = c.supportrepid
join invoice as i on i.customerid = c.customerid
group by e.employeeid"
```
Or in Python:
```python3
python3 - << 'EOF'
import sqlite3
conn = sqlite3.connect(":memory:")
conn.enable_load_extension(True)
conn.load_extension("build/zstd_vfs.so")
conn = sqlite3.connect("file:Chinook_Sqlite.zstd.sqlite?mode=ro&vfs=zstd", uri=True)
for row in conn.execute("""
select e.*, count(i.invoiceid) as 'Total Number of Sales'
from employee as e
join customer as c on e.employeeid = c.supportrepid
join invoice as i on i.customerid = c.customerid
group by e.employeeid
"""):
print(row)
EOF
```
Write to the compressed database:
```
sqlite3 :memory: -bail \
-cmd '.load build/zstd_vfs.so' \
-cmd '.open file:Chinook_Sqlite.zstd.sqlite?vfs=zstd' \
"update employee set Title = 'SVP Global Sales' where FirstName = 'Steve'"
```
Repeat the query to see the update.
The extension also *includes* [web_vfs](https://github.com/mlin/sqlite_web_vfs). A compressed database can be read from a HTTP(S) URL by opening the URI `file:/__web__?vfs=zstd&mode=ro&immutable=1&web_url={{PERCENT_ENCODED_URL}}`.
## Limitations
* Unix x86-64 oriented; help wanted for other platforms.
* [EXCLUSIVE locking mode](https://www.sqlite.org/pragma.html#pragma_locking_mode) always applies (a writer excludes all other connections for the lifetime of its own connection).
* Relaxing this is possible, but naturally demands rigorous concurrency testing. Help wanted.
* WAL mode isn't yet supported; do not touch.
* Several SQLite settings need tuning to get good performance (see below).
* Once more: **USE AT YOUR OWN RISK**
## Performance
Here are some operation timings using a [1,195MiB TPC-H database](https://github.com/lovasoa/TPCH-sqlite) on a Google N2 VM. This isn't a thorough benchmark, just a rough indication that many applications should find the de/compression overhead well worth the storage saved.
| | db file size | bulk load1 | Query 1 | Query 8 |
| -- | --: | --: | --: | --: |
| SQLite defaults | 1182MiB | 2.4s | 6.7s | 3.0s |
| zstd_vfs defaults | 647MiB | 25.0s | 8.8s | 35.7s |
| zstd_vfs tuned2 | 433MiB | 33.7s | 7.8s | 4.5s |
| zstd_vfs tuned &threads=8 | 433MiB | 6.9s | 6.7s | 3.1s |
1 by VACUUM INTO
2 `&level=6&outer_page_size=2048&outer_unsafe=true`; [`PRAGMA page_size=65536`](https://www.sqlite.org/pragma.html#pragma_page_size); [`PRAGMA cache_size=-102400`](https://www.sqlite.org/pragma.html#pragma_cache_size)
Query 1 is an aggregation satisfied by one table scan. Foreground Zstandard decompression slows it down by ~25% while the database file shrinks by 45-65%. (Each query starts with a hot filesystem cache and cold database page cache.)
Query 8 is an [historically influential](https://www.sqlite.org/queryplanner-ng.html) eight-way join. SQLite's default ~2MB page cache is too small for its access pattern, leading to a disastrous slowdown from repeated page decompression; but simply increasing the page cache to 100MiB largely solves this problem. Evidently, we should prefer a much larger page cache in view of the increased cost to miss.
Background threads can be used for both compression and "prefetching" during sequential scans. This greatly improves bulk load speed, and can sometimes fully hide decompression latency (given sequential access patterns, large pages, and spare CPU availability).
## Tuning
Some parameters are controlled from the file URI's query string opening the database, while others are set later through [PRAGMA statements](https://www.sqlite.org/pragma.html):
| | URI query parameters | PRAGMA |
| -- | -- | -- |
| writing/compression |
- level
- threads
- outer_page_size
- outer_unsafe
- page_size
- auto_vacuum
- journal_mode
| reading/decompression |
- outer_cache_size
- noprefetch
- cache_size
* **&level=3**: Zstandard compression level for newly written pages (-7 to 22)
* **&threads=1**: thread budget for page compression & prefetching (-1 to match host processors)
* **&outer_page_size=4096**: page size for the newly-created outer database; suggest doubling the (inner) page_size, to reduce space overhead from packing the compressed inner pages
* **&outer_unsafe=false**: set true to speed up bulk load by disabling transaction safety for outer database (app crash easily causes corruption)
* **&outer_cache_size=-2000**: page cache size for outer database, in [PRAGMA cache_size](https://www.sqlite.org/pragma.html#pragma_cache_size) units. Limited effect if on SSD.
* **&noprefetch=false**: set true to disable background prefetching/decompression even if threads>1. Prefetching is counterproductive if page_size is too small or CPU cycles are scarce.
* **PRAGMA page_size=4096**: uncompressed [page size](https://www.sqlite.org/pragma.html#pragma_page_size) for the newly-created inner database. Larger pages are more compressible, but increase [read/write amplification](http://smalldatum.blogspot.com/2015/11/read-write-space-amplification-pick-2_23.html). YMMV but 8 or 16 KiB have been working well for us.
* **PRAGMA auto_vacuum=NONE**: set to FULL or INCREMENTAL on a newly-created database if you expect its size to fluctuate over time, so that the file will [shrink to fit](https://www.sqlite.org/pragma.html#pragma_auto_vacuum). (The outer database auto-vacuums when it's closed.)
* **PRAGMA journal_mode=DELETE**: set to MEMORY or OFF as discussed in [ZIPVFS docs](https://www.sqlite.org/zipvfs/doc/trunk/www/howitworks.wiki) *Multple Journal Files*. (As explained there, this shouldn't compromise transaction safety.)
* **PRAGMA cache_size=-2000**: page cache size for inner database, in [PRAGMA cache_size](https://www.sqlite.org/pragma.html#pragma_cache_size) units. Critical for complex query performance, as illustrated above.
For bulk load, open the database with the `&outer_unsafe=true` URI parameter and `SQLITE_OPEN_NOMUTEX` flag, then issue `PRAGMA journal_mode=OFF; PRAGMA synchronous=OFF`. Insert everything within one big transaction, create any needed indices, and commit.
`VACUUM INTO 'file:fresh_copy.db?vfs=zstd'` (+ URI parameters above) is a suggested way to [defragment](https://www.sqlite.org/lang_vacuum.html) a database that's been heavily modified over time.