https://github.com/mrpowers/chispa
PySpark test helper methods with beautiful error messages
https://github.com/mrpowers/chispa
pyspark testing
Last synced: 8 days ago
JSON representation
PySpark test helper methods with beautiful error messages
- Host: GitHub
- URL: https://github.com/mrpowers/chispa
- Owner: MrPowers
- License: mit
- Created: 2019-03-19T15:52:21.000Z (about 6 years ago)
- Default Branch: main
- Last Pushed: 2025-03-31T12:03:40.000Z (18 days ago)
- Last Synced: 2025-04-03T13:05:57.633Z (15 days ago)
- Topics: pyspark, testing
- Language: Python
- Homepage: https://mrpowers.github.io/chispa/
- Size: 3.63 MB
- Stars: 681
- Watchers: 4
- Forks: 71
- Open Issues: 43
-
Metadata Files:
- Readme: README.md
- Contributing: CONTRIBUTING.md
- License: LICENSE.md
Awesome Lists containing this project
- awesome-databricks - chispa - commit/mrpowers/chispa.svg"> - PySpark test helper methods with beautiful error messages (External Resources / Repos)
README
# chispa
](https://github.com/MrPowers/chispa/actions/workflows/ci.yml/badge.svg)

[](https://badge.fury.io/py/chispa)
chispa provides fast PySpark test helper methods that output descriptive error messages.
This library makes it easy to write high quality PySpark code.
Fun fact: "chispa" means Spark in Spanish ;)
## Installation
Install the latest version with `pip install chispa`.
If you use Poetry, add this library as a development dependency with `poetry add chispa -G dev`.
## Column equality
Suppose you have a function that removes the non-word characters in a string.
```python
def remove_non_word_characters(col):
return F.regexp_replace(col, "[^\\w\\s]+", "")
```
Create a `SparkSession` so you can create DataFrames.
```python
from pyspark.sql import SparkSession
spark = (SparkSession.builder
.master("local")
.appName("chispa")
.getOrCreate())
```
Create a DataFrame with a column that contains strings with non-word characters, run the `remove_non_word_characters` function, and check that all these characters are removed with the chispa `assert_column_equality` method.
```python
import pytest
from chispa.column_comparer import assert_column_equality
import pyspark.sql.functions as F
def test_remove_non_word_characters_short():
data = [
("jo&&se", "jose"),
("**li**", "li"),
("#::luisa", "luisa"),
(None, None)
]
df = (spark.createDataFrame(data, ["name", "expected_name"])
.withColumn("clean_name", remove_non_word_characters(F.col("name"))))
assert_column_equality(df, "clean_name", "expected_name")
```
Let's write another test that'll fail to see how the descriptive error message lets you easily debug the underlying issue.
Here's the failing test:
```python
def test_remove_non_word_characters_nice_error():
data = [
("matt7", "matt"),
("bill&", "bill"),
("isabela*", "isabela"),
(None, None)
]
df = (spark.createDataFrame(data, ["name", "expected_name"])
.withColumn("clean_name", remove_non_word_characters(F.col("name"))))
assert_column_equality(df, "clean_name", "expected_name")
```
Here's the nicely formatted error message:
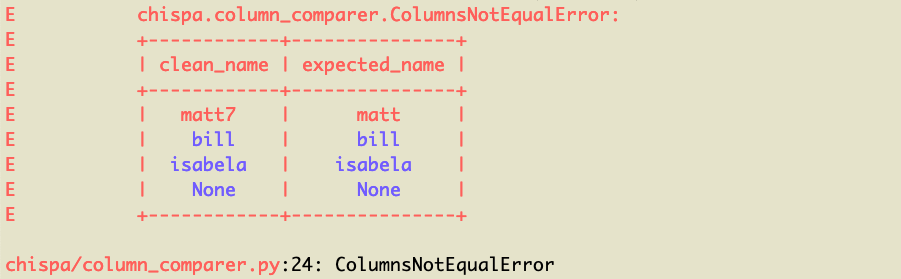
You can see the `matt7` / `matt` row of data is what's causing the error (note it's highlighted in red). The other rows are colored blue because they're equal.
## DataFrame equality
We can also test the `remove_non_word_characters` method by creating two DataFrames and verifying that they're equal.
Creating two DataFrames is slower and requires more code, but comparing entire DataFrames is necessary for some tests.
```python
from chispa.dataframe_comparer import *
def test_remove_non_word_characters_long():
source_data = [
("jo&&se",),
("**li**",),
("#::luisa",),
(None,)
]
source_df = spark.createDataFrame(source_data, ["name"])
actual_df = source_df.withColumn(
"clean_name",
remove_non_word_characters(F.col("name"))
)
expected_data = [
("jo&&se", "jose"),
("**li**", "li"),
("#::luisa", "luisa"),
(None, None)
]
expected_df = spark.createDataFrame(expected_data, ["name", "clean_name"])
assert_df_equality(actual_df, expected_df)
```
Let's write another test that'll return an error, so you can see the descriptive error message.
```python
def test_remove_non_word_characters_long_error():
source_data = [
("matt7",),
("bill&",),
("isabela*",),
(None,)
]
source_df = spark.createDataFrame(source_data, ["name"])
actual_df = source_df.withColumn(
"clean_name",
remove_non_word_characters(F.col("name"))
)
expected_data = [
("matt7", "matt"),
("bill&", "bill"),
("isabela*", "isabela"),
(None, None)
]
expected_df = spark.createDataFrame(expected_data, ["name", "clean_name"])
assert_df_equality(actual_df, expected_df)
```
Here's the nicely formatted error message:

### Ignore row order
You can easily compare DataFrames, ignoring the order of the rows. The content of the DataFrames is usually what matters, not the order of the rows.
Here are the contents of `df1`:
```
+--------+
|some_num|
+--------+
| 1|
| 2|
| 3|
+--------+
```
Here are the contents of `df2`:
```
+--------+
|some_num|
+--------+
| 2|
| 1|
| 3|
+--------+
```
Here's how to confirm `df1` and `df2` are equal when the row order is ignored.
```python
assert_df_equality(df1, df2, ignore_row_order=True)
```
If you don't specify to `ignore_row_order` then the test will error out with this message:
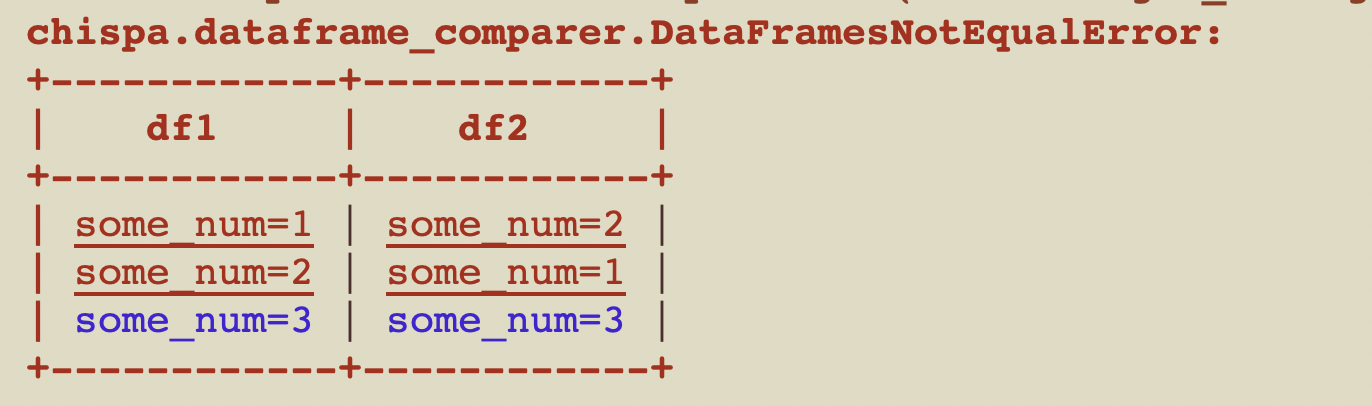
The rows aren't ordered by default because sorting slows down the function.
### Ignore column order
This section explains how to compare DataFrames, ignoring the order of the columns.
Suppose you have the following `df1`:
```
+----+----+
|num1|num2|
+----+----+
| 1| 7|
| 2| 8|
| 3| 9|
+----+----+
```
Here are the contents of `df2`:
```
+----+----+
|num2|num1|
+----+----+
| 7| 1|
| 8| 2|
| 9| 3|
+----+----+
```
Here's how to compare the equality of `df1` and `df2`, ignoring the column order:
```python
assert_df_equality(df1, df2, ignore_column_order=True)
```
Here's the error message you'll see if you run `assert_df_equality(df1, df2)`, without ignoring the column order.

### Ignore specific columns
This section explains how to compare DataFrames, ignoring specific columns.
Suppose you have the following `df1`:
```
+------------+-------------+
| name | clean_name |
+------------+-------------+
| "matt7" | "matt7" |
| "bill&" | "bill" |
| "isabela*" | "isabela" |
| "None" | "None" |
+------------+-------------+
```
Here are the contents of `df2`:
```
+------------+-------------+
| name | clean_name |
+------------+-------------+
| "matt7" | "matt" |
| "bill&" | "bill" |
| "isabela*" | "isabela" |
| "None" | "None" |
+------------+-------------+
```
Here's how to compare the equality of `df1` and `df2`, ignoring the column `clean_name`:
```python
assert_df_equality(df1, df2, ignore_columns=["clean_name"])
```
Here's the error message you'll see if you run `assert_df_equality(df1, df2)`, without ignoring the column `clean_name`.

### Ignore nullability
Each column in a schema has three properties: a name, data type, and nullable property. The column can accept null values if `nullable` is set to true.
You'll sometimes want to ignore the nullable property when making DataFrame comparisons.
Suppose you have the following `df1`:
```
+-----+---+
| name|age|
+-----+---+
| juan| 7|
|bruna| 8|
+-----+---+
```
And this `df2`:
```
+-----+---+
| name|age|
+-----+---+
| juan| 7|
|bruna| 8|
+-----+---+
```
You might be surprised to find that in this example, `df1` and `df2` are not equal and will error out with this message:

Examine the code in this contrived example to better understand the error:
```python
def ignore_nullable_property():
s1 = StructType([
StructField("name", StringType(), True),
StructField("age", IntegerType(), True)])
df1 = spark.createDataFrame([("juan", 7), ("bruna", 8)], s1)
s2 = StructType([
StructField("name", StringType(), True),
StructField("age", IntegerType(), False)])
df2 = spark.createDataFrame([("juan", 7), ("bruna", 8)], s2)
assert_df_equality(df1, df2)
```
You can ignore the nullable property when assessing equality by adding a flag:
```python
assert_df_equality(df1, df2, ignore_nullable=True)
```
Elements contained within an `ArrayType()` also have a nullable property, in addition to the nullable property of the column schema. These are also ignored when passing `ignore_nullable=True`.
Again, examine the following code to understand the error that `ignore_nullable=True` bypasses:
```python
def ignore_nullable_property_array():
s1 = StructType([
StructField("name", StringType(), True),
StructField("coords", ArrayType(DoubleType(), True), True),])
df1 = spark.createDataFrame([("juan", [1.42, 3.5]), ("bruna", [2.76, 3.2])], s1)
s2 = StructType([
StructField("name", StringType(), True),
StructField("coords", ArrayType(DoubleType(), False), True),])
df2 = spark.createDataFrame([("juan", [1.42, 3.5]), ("bruna", [2.76, 3.2])], s2)
assert_df_equality(df1, df2)
```
### Allow NaN equality
Python has NaN (not a number) values and two NaN values are not considered equal by default. Create two NaN values, compare them, and confirm they're not considered equal by default.
```python
nan1 = float('nan')
nan2 = float('nan')
nan1 == nan2 # False
```
pandas considers NaN values to be equal by default, but this library requires you to set a flag to consider two NaN values to be equal.
```python
assert_df_equality(df1, df2, allow_nan_equality=True)
```
## Customize formatting
You can specify custom formats for the printed error messages as follows:
```python
from chispa import FormattingConfig
formats = FormattingConfig(
mismatched_rows={"color": "light_yellow"},
matched_rows={"color": "cyan", "style": "bold"},
mismatched_cells={"color": "purple"},
matched_cells={"color": "blue"},
)
assert_basic_rows_equality(df1.collect(), df2.collect(), formats=formats)
```
or similarly:
```python
from chispa import FormattingConfig, Color, Style
formats = FormattingConfig(
mismatched_rows={"color": Color.LIGHT_YELLOW},
matched_rows={"color": Color.CYAN, "style": Style.BOLD},
mismatched_cells={"color": Color.PURPLE},
matched_cells={"color": Color.BLUE},
)
assert_basic_rows_equality(df1.collect(), df2.collect(), formats=formats)
```
You can also define these formats in `conftest.py` and inject them via a fixture:
```python
@pytest.fixture()
def chispa_formats():
return FormattingConfig(
mismatched_rows={"color": "light_yellow"},
matched_rows={"color": "cyan", "style": "bold"},
mismatched_cells={"color": "purple"},
matched_cells={"color": "blue"},
)
def test_shows_assert_basic_rows_equality(chispa_formats):
...
assert_basic_rows_equality(df1.collect(), df2.collect(), formats=chispa_formats)
```
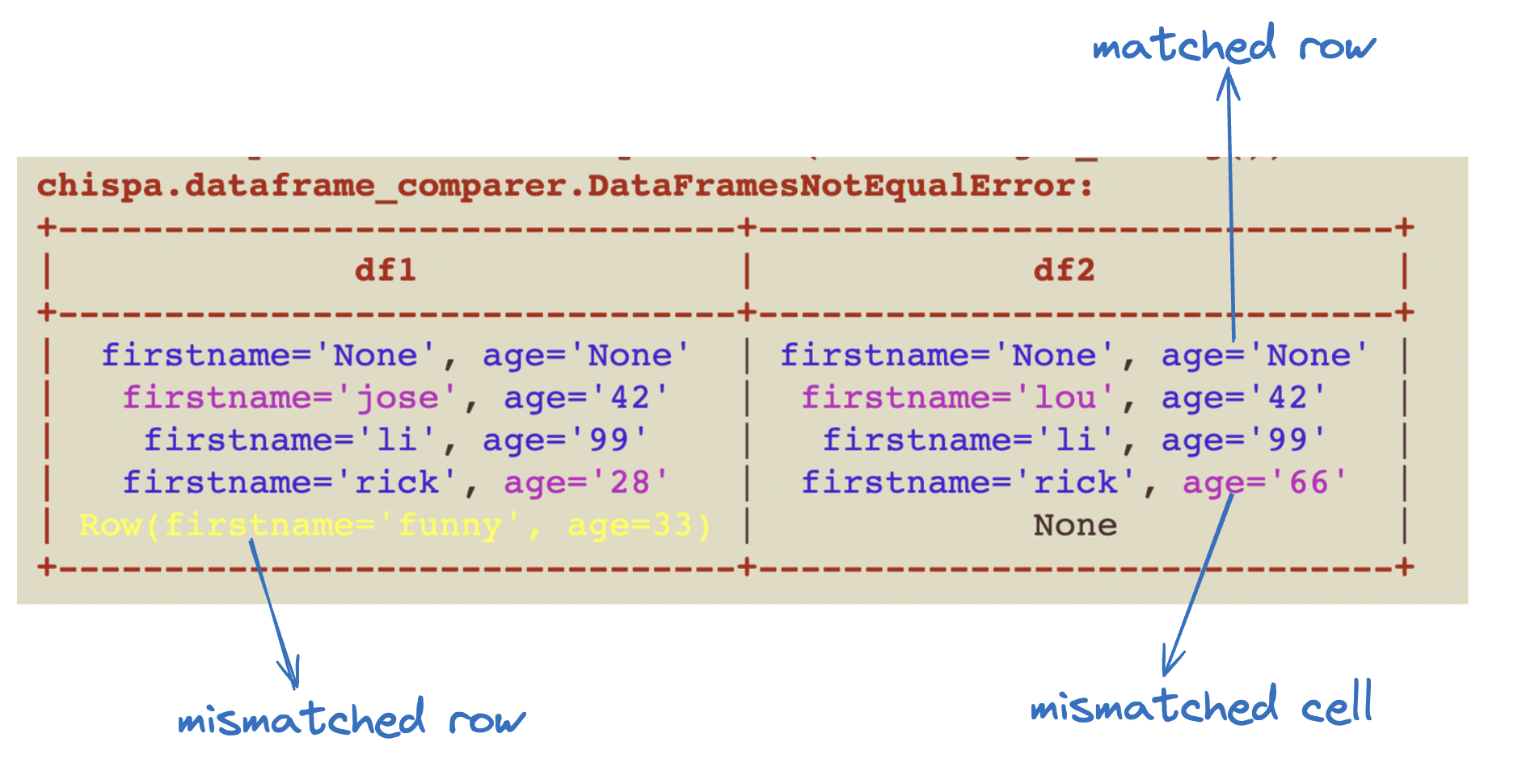
## Approximate column equality
We can check if columns are approximately equal, which is especially useful for floating number comparisons.
Here's a test that creates a DataFrame with two floating point columns and verifies that the columns are approximately equal. In this example, values are considered approximately equal if the difference is less than 0.1.
```python
def test_approx_col_equality_same():
data = [
(1.1, 1.1),
(2.2, 2.15),
(3.3, 3.37),
(None, None)
]
df = spark.createDataFrame(data, ["num1", "num2"])
assert_approx_column_equality(df, "num1", "num2", 0.1)
```
Here's an example of a test with columns that are not approximately equal.
```python
def test_approx_col_equality_different():
data = [
(1.1, 1.1),
(2.2, 2.15),
(3.3, 5.0),
(None, None)
]
df = spark.createDataFrame(data, ["num1", "num2"])
assert_approx_column_equality(df, "num1", "num2", 0.1)
```
This failing test will output a readable error message so the issue is easy to debug.

## Approximate DataFrame equality
Let's create two DataFrames and confirm they're approximately equal.
```python
def test_approx_df_equality_same():
data1 = [
(1.1, "a"),
(2.2, "b"),
(3.3, "c"),
(None, None)
]
df1 = spark.createDataFrame(data1, ["num", "letter"])
data2 = [
(1.05, "a"),
(2.13, "b"),
(3.3, "c"),
(None, None)
]
df2 = spark.createDataFrame(data2, ["num", "letter"])
assert_approx_df_equality(df1, df2, 0.1)
```
The `assert_approx_df_equality` method is smart and will only perform approximate equality operations for floating point numbers in DataFrames. It'll perform regular equality for strings and other types.
Let's perform an approximate equality comparison for two DataFrames that are not equal.
```python
def test_approx_df_equality_different():
data1 = [
(1.1, "a"),
(2.2, "b"),
(3.3, "c"),
(None, None)
]
df1 = spark.createDataFrame(data1, ["num", "letter"])
data2 = [
(1.1, "a"),
(5.0, "b"),
(3.3, "z"),
(None, None)
]
df2 = spark.createDataFrame(data2, ["num", "letter"])
assert_approx_df_equality(df1, df2, 0.1)
```
Here's the pretty error message that's outputted:
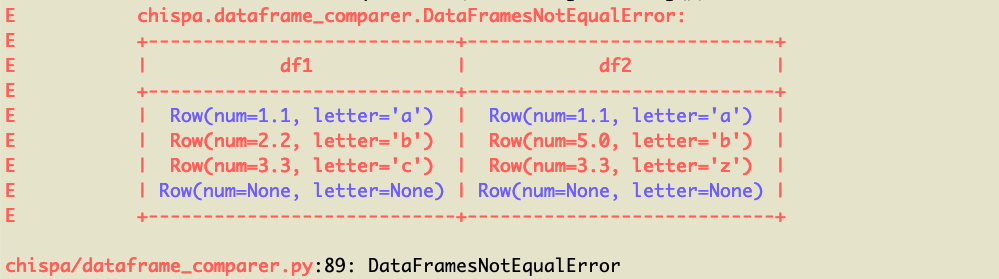
## Schema mismatch messages
DataFrame equality messages peform schema comparisons before analyzing the actual content of the DataFrames. DataFrames that don't have the same schemas should error out as fast as possible.
Let's compare a DataFrame that has a string column an integer column with a DataFrame that has two integer columns to observe the schema mismatch message.
```python
def test_schema_mismatch_message():
data1 = [
(1, "a"),
(2, "b"),
(3, "c"),
(None, None)
]
df1 = spark.createDataFrame(data1, ["num", "letter"])
data2 = [
(1, 6),
(2, 7),
(3, 8),
(None, None)
]
df2 = spark.createDataFrame(data2, ["num", "num2"])
assert_df_equality(df1, df2)
```
Here's the error message:

## Supported PySpark / Python versions
chispa currently supports PySpark 2.4+ and Python 3.5+.
Use chispa v0.8.2 if you're using an older Python version.
PySpark 2 support will be dropped when chispa 1.x is released.
## Benchmarks
TODO: Need to benchmark these methods vs. the spark-testing-base ones
## Developing chispa on your local machine
You are encouraged to clone and/or fork this repo.
This project uses [Poetry](https://python-poetry.org/) for packaging and dependency management.
* Setup the virtual environment with `poetry install`
* Run the tests with `poetry run pytest tests`
Studying the codebase is a great way to learn about PySpark!
## Contributing
Anyone is encouraged to submit a pull request, open an issue, or submit a bug report.
We're happy to promote folks to be library maintainers if they make good contributions.