https://github.com/mrtarantoga/mdl-density-histogram
Cython-accelerated MDL histogram density estimation. Implements Kontkanen & Myllymaki's dynamic programming algorithm for optimal variable-width bins, parametric complexity with Ramanujan approximations, and automatic bin count selection. Based on "MDL Histogram Density Estimation" (JMLR 2007).
https://github.com/mrtarantoga/mdl-density-histogram
cython-library mdl numpy pypi-package python3
Last synced: 2 months ago
JSON representation
Cython-accelerated MDL histogram density estimation. Implements Kontkanen & Myllymaki's dynamic programming algorithm for optimal variable-width bins, parametric complexity with Ramanujan approximations, and automatic bin count selection. Based on "MDL Histogram Density Estimation" (JMLR 2007).
- Host: GitHub
- URL: https://github.com/mrtarantoga/mdl-density-histogram
- Owner: MrTarantoga
- License: apache-2.0
- Created: 2025-07-30T13:07:03.000Z (6 months ago)
- Default Branch: main
- Last Pushed: 2025-09-18T12:19:59.000Z (4 months ago)
- Last Synced: 2025-09-26T04:57:02.300Z (4 months ago)
- Topics: cython-library, mdl, numpy, pypi-package, python3
- Language: Python
- Homepage: https://pypi.org/project/MDL-Density-Histogram/
- Size: 125 KB
- Stars: 1
- Watchers: 0
- Forks: 0
- Open Issues: 0
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
README
[](https://github.com/MrTarantoga/MDL-Density-Histogram/actions/workflows/python-publish.yml)
[](https://github.com/MrTarantoga/MDL-Density-Histogram/actions/workflows/python-app.yml)
# MDL Optimal Histogram Density Estimation
This package provides a Cython-accelerated implementation of the **Minimum Description Length (MDL) optimal histogram density estimation** algorithm from Kontkanen & Myllymaki (2007). It uses information-theoretic principles to automatically determine optimal variable-width bins for density estimation.
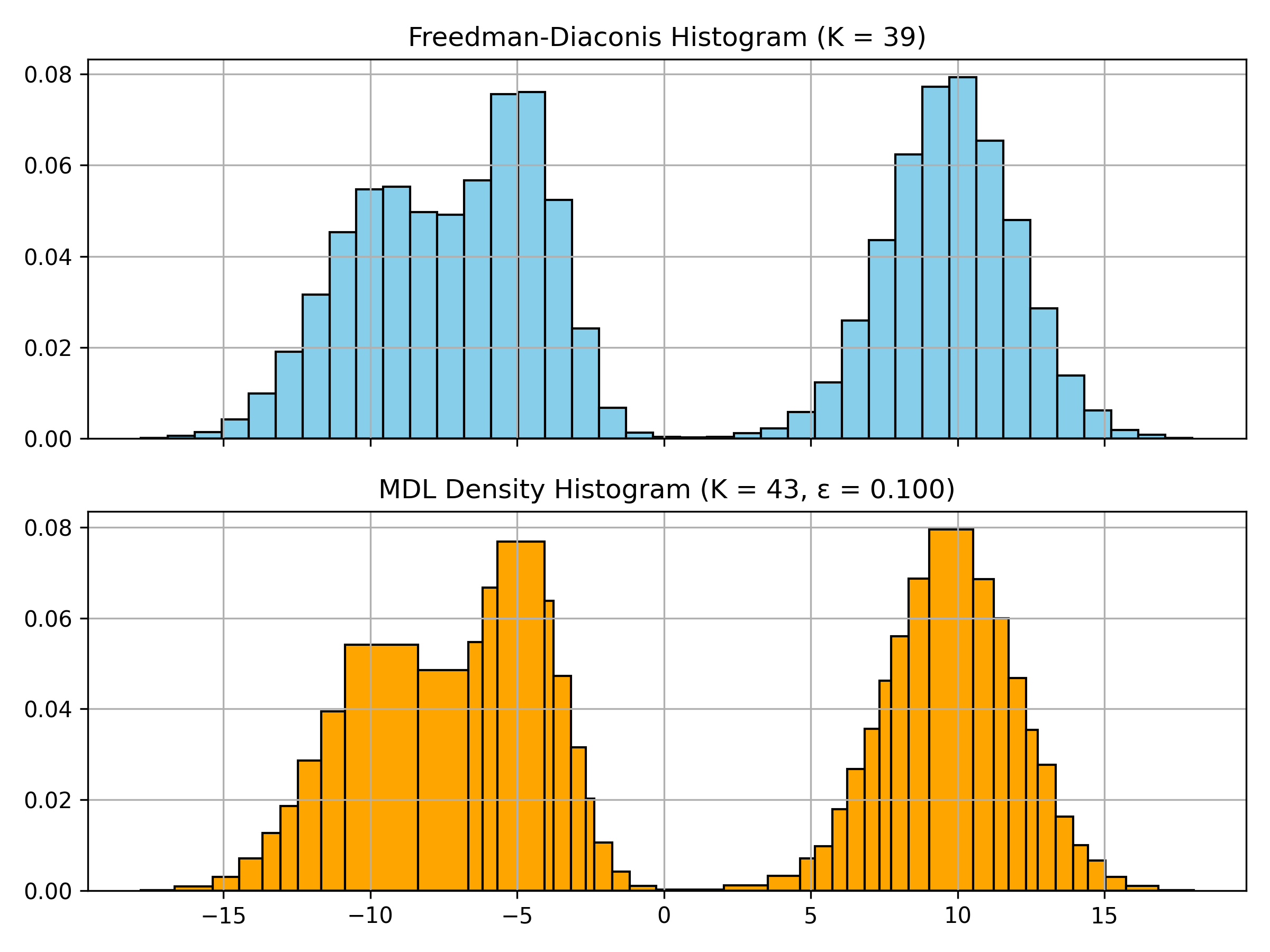
## Features
- **MDL Principle**: Uses stochastic complexity for model selection
- **Dynamic Programming**: Efficient O(E²·K_max) optimization (cache parametric complexity computation, speed up)
- **Score of each *K*th bin**: The score of each bin is returned to understand the performance of different properties of the same dataset.
- **Variable-Width Bins**: Adapts to data density variations
- **Automatic Bin Count**: No manual parameter tuning required (except maximum bin count to consider $K_{max}$ and data resolution $\epsilon$)
- **Cython Acceleration**: Critical operations compiled to C
## Installation
You can install the package using pip:
```bash
pip install MDL-Density-Histogram
```
Alternatively, you can install it from source by cloning the repository and running:
```bash
# From project root directory
pip install .
```
Requires:
- Python 3.11+
- NumPy
- Cython
- C compiler (GCC/Clang/MSVC)
## Usage Example
```python
import numpy as np
from mdl_density_hist import mdl_optimal_histogram
# Generate sample data
data = np.random.normal(0, 1, 1000)
# Compute optimal histogram
cut_points, K_scores = mdl_optimal_histogram(data, epsilon=0.1)
# Print score of each bin
print(f"K_scores: {K_scores}")
# Visualize result
import matplotlib.pyplot as plt
plt.hist(data, bins=cut_points, density=True)
plt.title('MDL Optimal Histogram')
plt.show()
```
## Parameters
- `data`: Input array (1D numpy array)
- `epsilon`: Quantization precision (default: 0.1)
- `K_max`: Maximum number of bins (default: 10)
## Algorithm Highlights
- Uses **Ramanujan's factorial approximation** for efficient parametric complexity
- Cache parameteric complexity to speed up computation
## Paper Citation
Kontkanen, P., & Myllymäki, P. (2007).
*MDL Histogram Density Estimation*
Journal of Machine Learning Research 8 (2007)
[PDF](https://proceedings.mlr.press/v2/kontkanen07a/kontkanen07a.pdf)
## License
Apache 2.0 License - See LICENSE file
## Project Structure
```
src/
├── mdl_density_hist/
│ ├── __init__.py
│ └── mdl_hist.pyx # Core Cython implementation
└── pyproject.toml
```
## Performance Notes
- Precomputed parametric complexity using dynamic programming
- Memory-optimized array operations via NumPy
- Candidate cut point pruning for reduced search space
For implementation details, see the [paper](https://proceedings.mlr.press/v2/kontkanen07a/kontkanen07a.pdf) and inline code comments.