https://github.com/multiomics-analytics-group/abaco
Batch Effect Correction framework for metagenomic data
https://github.com/multiomics-analytics-group/abaco
batch-correction bioinformatics-software metagenomics microbiome python
Last synced: about 1 month ago
JSON representation
Batch Effect Correction framework for metagenomic data
- Host: GitHub
- URL: https://github.com/multiomics-analytics-group/abaco
- Owner: Multiomics-Analytics-Group
- License: other
- Created: 2025-07-22T11:29:51.000Z (3 months ago)
- Default Branch: main
- Last Pushed: 2025-08-25T12:15:36.000Z (about 1 month ago)
- Last Synced: 2025-08-25T14:26:19.082Z (about 1 month ago)
- Topics: batch-correction, bioinformatics-software, metagenomics, microbiome, python
- Language: Python
- Homepage: https://mona-abaco.readthedocs.io/
- Size: 6.54 MB
- Stars: 2
- Watchers: 0
- Forks: 0
- Open Issues: 0
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
README
## 
Batch Effect Correction framework for metagenomic data
## Table of Contents
- [About the project](#about-the-project)
- [Installation](#installation)
- [Features](#features)
- [Usage](#usage)
- [Documentation](#documentation)
- [License](#license)
- [Contributing](#contributing)
- [Credits and acknowledgements](#credits-and-acknowledgements)
- [Contact and feedback](#contact-and-feedback)
## About the project
The integration of metagenomic data from multiple studies and experimental conditions is essential to understand the interactions between microbial communities in complex biological systems, but the inherent diversity and biological complexity pose methodological challenges that require refined strategies for atlas-level integration. ABaCo, a family of generative models based on Variational Autoencoders (VAEs) combined with an adversarial training, aim for the integration of metagenomic data from different studies by minimizing technical heterogeneity conserving biological significance. The VAE encodes the data into a latent space, while the discriminator is trained to detect the provenance of the data, eliminating variability associated with its origin; concurrently, the data is modeled using distributions suitable for raw counts, and the latent space follows a clustering prior to ensure biological conservation.
An overview of the ABaCo workflow is shown in the figure below:
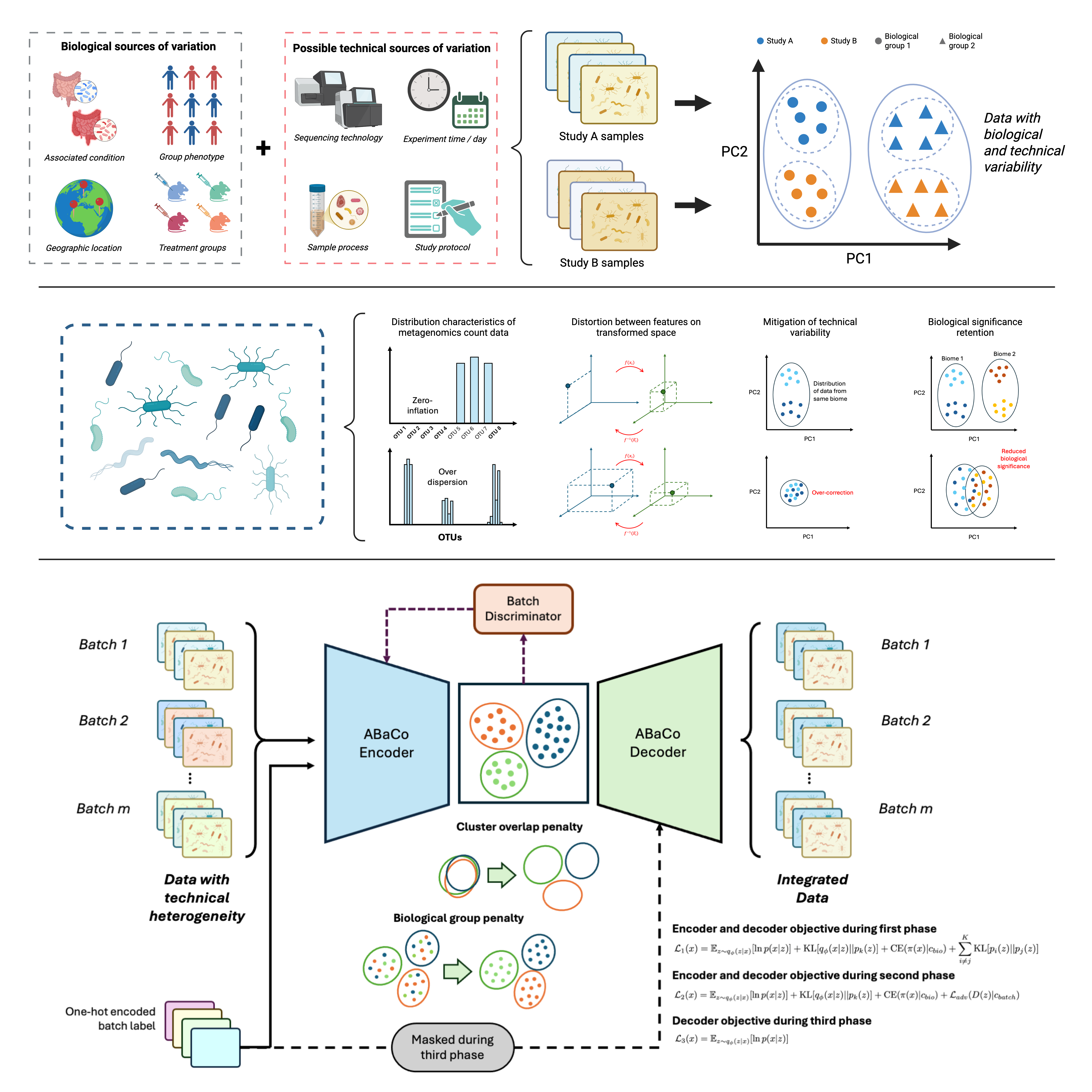
## Installation
> [!TIP]
> It is recommended to install ABaCo inside a virtual environment to manage depenendencies and avoid conflicts with existing packages. You can use the virtual environment manager of your choice, such as `poetry`, `conda`, or `pipenv`.
### Pip
ABaCo is available on [PyPI][abaco-pypi] and can be installed using pip:
```bash
pip install abaco
```
You can also install the package for development by cloning this repository and running the following command:
> [!WARNING]
> We assume you are in the root directory of the cloned repository when running this command. Otherwise, you need to specify the path to the `abaco` directory.
```bash
pip install -e .
```
## Features
## Usage
## Documentation
ABaCo's documentation is hosted on [Read the Docs][abaco-docs]. It includes detailed examples, configuration options, and the API reference.
## License
The code in this repository is licensed under the **MIT License**, allowing you to use, modify, and distribute it freely as long as you include the original copyright and license notice.
The documentation and other creative content are licensed under the **Creative Commons Attribution 4.0 International (CC BY 4.0) License**, meaning you are free to share and adapt it with proper attribution.
Full details for both licenses can be found in the [LICENSE][abaco-license] file.
## Contributing
ABaCo is an open-source project, and we welcome contributions of all kinds via GitHub issues and pull requests. You can report bugs, suggest improvements, propose new features, or implement changes. Please follow the guidelines in the [CONTRIBUTING](CONTRIBUTING.md) file to ensure that your contribution is easily integrated into the project.
## Credits and acknowledgements
- ABaCo was developed by the [Multiomics Network Analytics Group (MoNA)][Mona] at the [Novo Nordisk Foundation Center for Biosustainability (DTU Biosustain)][Biosustain].
## Contact and feedback
We appreciate your feedback! If you have any comments, suggestions, or run into issues while using ABaCo, feel free to [open an issue][new-issue] in this repository. Your input helps us make ABaCo better for everyone.
[abaco-pypi]: https://pypi.org/project/abaco/
[abaco-license]: https://github.com/Multiomics-Analytics-Group/abaco/blob/main/LICENSE.md
[abaco-docs]: https://mona-abaco.readthedocs.io/
[Mona]: https://multiomics-analytics-group.github.io/
[Biosustain]: https://www.biosustain.dtu.dk/
[new-issue]:https://github.com/Multiomics-Analytics-Group/abaco/issues/new





