https://github.com/multiomics-analytics-group/vuegen
VueGen automates the creation of reports from bioinformatics outputs, supporting formats like PDF, HTML, DOCX, ODT, PPTX, Reveal.js, Jupyter notebooks, and Streamlit web applications. Users simply provide a directory with output files and VueGen compiles them into a structured report.
https://github.com/multiomics-analytics-group/vuegen
data-visualization developer-tools python quarto reports streamlit
Last synced: 7 months ago
JSON representation
VueGen automates the creation of reports from bioinformatics outputs, supporting formats like PDF, HTML, DOCX, ODT, PPTX, Reveal.js, Jupyter notebooks, and Streamlit web applications. Users simply provide a directory with output files and VueGen compiles them into a structured report.
- Host: GitHub
- URL: https://github.com/multiomics-analytics-group/vuegen
- Owner: Multiomics-Analytics-Group
- License: mit
- Created: 2023-06-13T09:09:35.000Z (over 2 years ago)
- Default Branch: main
- Last Pushed: 2025-03-18T08:19:29.000Z (7 months ago)
- Last Synced: 2025-03-18T09:29:36.777Z (7 months ago)
- Topics: data-visualization, developer-tools, python, quarto, reports, streamlit
- Language: Python
- Homepage: https://vuegen.readthedocs.io
- Size: 68.8 MB
- Stars: 10
- Watchers: 2
- Forks: 0
- Open Issues: 24
-
Metadata Files:
- Readme: README.md
- Contributing: CONTRIBUTING.md
- License: LICENSE
- Citation: CITATION.cff
Awesome Lists containing this project
README

-----------------
VueGen is a Python package that automates the creation of scientific reports.
| Information | Links |
| :--- | :--- |
| **Package** |[][vuegen-pypi] [][vuegen-conda] [][vuegen-pypi] [][vuegen-docker-quay] [][vuegen-license]|
| **Documentation** | [](https://www.sphinx-doc.org/)  [][vuegen-docs]|
| **Build** | [][ci-gh-action] [][ci-docs]|
| **Examples** | [][emp-html-demo] [][emp-st-demo]|
| **Discuss on GitHub** | [][issues] [][pulls]|
| **Cite** | [][vuegen-preprint]|
## Table of contents:
- [About the project](#about-the-project)
- [Installation](#installation)
- [Execution](#execution)
- [Case studies](#case-studies)
- [Web application deployment](#web-application-deployment)
- [Contributing](#contributing)
- [Credits and acknowledgements](#credits-and-acknowledgements)
- [Citation](#citation)
- [Contact and feedback](#contact-and-feedback)
## About the project
VueGen automates the creation of reports based on a directory with plots, dataframes, and other files in different formats. A YAML configuration file is generated from the directory to define the structure of the report. Users can customize the report by modifying the configuration file, or they can create their own configuration file instead of passing a directory as input.
The configuration file specifies the structure of the report, including sections, subsections, and various components such as plots, dataframes, markdown, html, and API calls. Reports can be generated in various formats, including documents (PDF, HTML, DOCX, ODT), presentations (PPTX, Reveal.js), notebooks (Jupyter) or [Streamlit](streamlit) web applications.
An overview of the VueGen workflow is shown in the figure below:
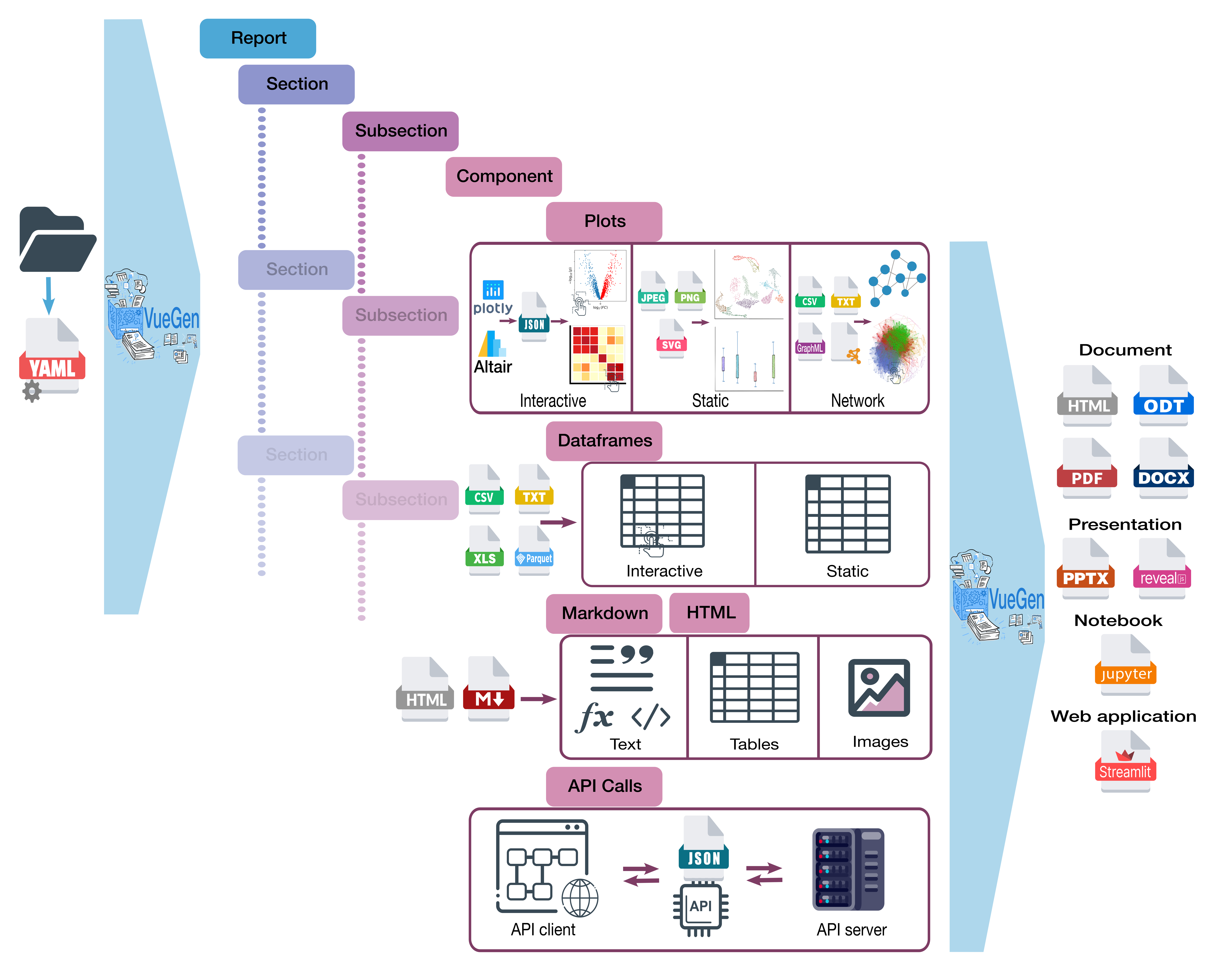
Also, the class diagram for the project is presented below to illustrate the architecture and relationships between classes:
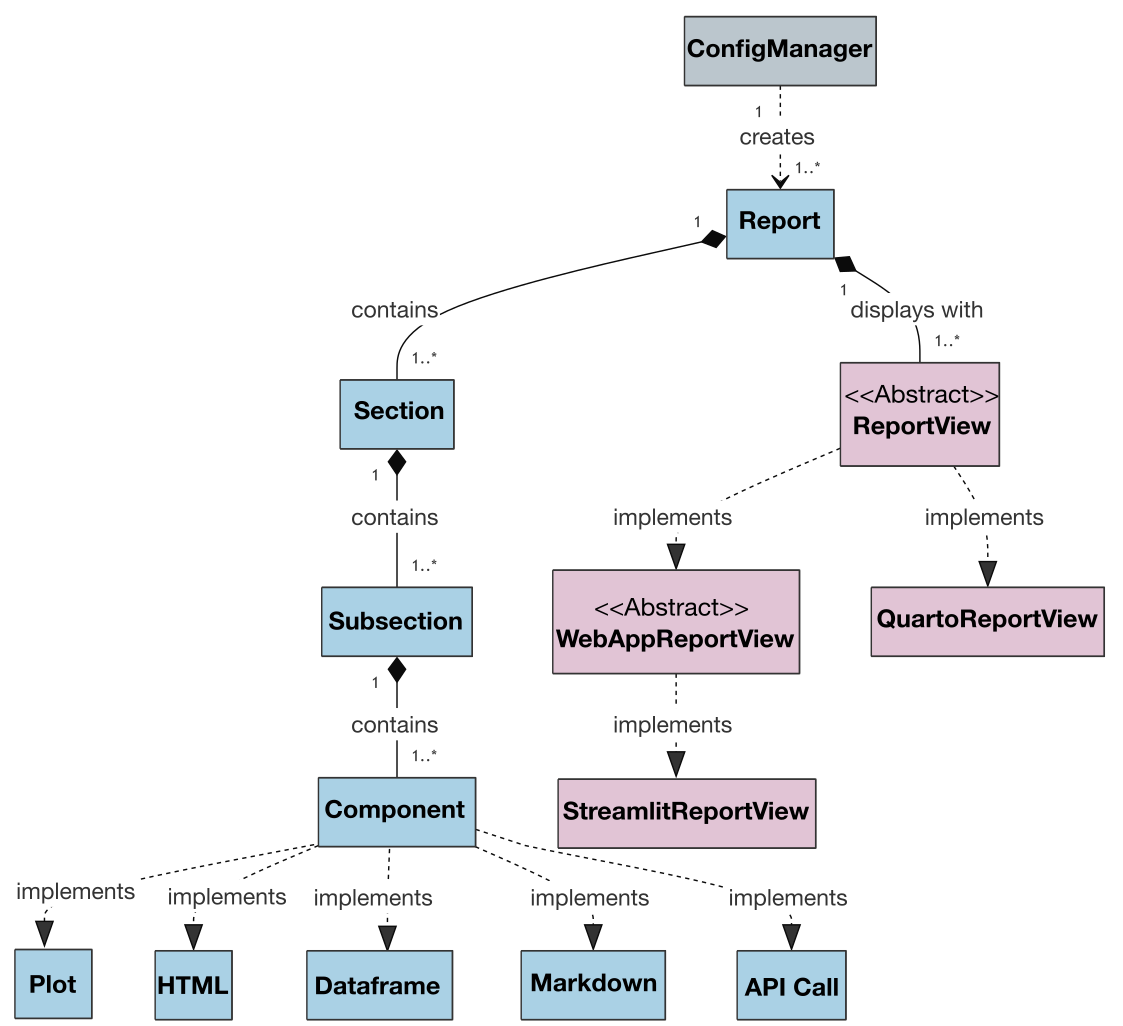
An extended version of the class diagram with attributes and methods is available [here][vuegen-class-diag-att].
The VueGen documentation is available at [vuegen.readthedocs.io][vuegen-docs], where you can find detailed information of the package’s classes and functions, installation and execution instructions, and case studies to demonstrate its functionality.
## Installation
> [!TIP]
> It is recommended to install VueGen inside a virtual environment to manage depenendencies and avoid conflicts with existing packages. You can use the virtual environment manager of your choice, such as `poetry`, `conda`, or `pipenv`.
### Pip
VueGen is available on [PyPI][vuegen-pypi] and can be installed using pip:
```bash
pip install vuegen
```
You can also install the package for development by cloning this repository and running the following command:
```bash
pip install -e path/to/vuegen # specify location
pip install -e . # in case your pwd is in the vuegen directory
```
### Conda
VueGen is also available on [Bioconda][vuegen-conda] and can be installed using conda:
```bash
conda install bioconda::vuegen
```
### Dependencies
VueGen uses [Quarto][quarto] to generate various report types. The pip insallation includes quarto using the [quarto-cli Python library][quarto-cli-pypi]. To test if quarto is installed in your computer, run the following command:
```bash
quarto check
```
> [!TIP]
> If quarto is not installed, you can download the command-line interface from the [Quarto website][quarto-cli] for your operating system.
### Docker
If you prefer not to install VueGen on your system, a pre-configured Docker container is available. It includes all dependencies, ensuring a fully reproducible execution environment. See the [Execution section](#execution) for details on running VueGen with Docker. The official Docker image is available at [quay.io/dtu_biosustain_dsp/vuegen][vuegen-docker-quay].
### Nextflow and nf-core
VueGen is also available as a [nf-core][nfcore] module, customised for compatibility with the [Nextflow][nextflow] environment. This module is designed to automate report generation from outputs produced by other modules, subworkflows, or pipelines. The code and documentation for the nf-core module are available in the [nf-VueGen repository][nf-vuegen].
## Execution
> [!IMPORTANT]
> Here we use the `Earth_microbiome_vuegen_demo_notebook` directory and the `Earth_microbiome_vuegen_demo_notebook.yaml` configuration file as examples, which are available in the `docs/example_data` and `docs/example_config_files` folders, respectively. Make sure to clone this reposiotry to access these contents, or use your own directory and configuration file.
Run VueGen using a directory with the following command:
```bash
vuegen --directory docs/example_data/Earth_microbiome_vuegen_demo_notebook --report_type streamlit
```
> [!NOTE]
> By default, the `streamlit_autorun` argument is set to False, but you can use it in case you want to automatically run the streamlit app.
It's also possible to provide a configuration file instead of a directory:
```bash
vuegen --config docs/example_config_files/Earth_microbiome_vuegen_demo_notebook.yaml --report_type streamlit
```
The current report types supported by VueGen are:
* Streamlit
* HTML
* PDF
* DOCX
* ODT
* Reveal.js
* PPTX
* Jupyter
### Running VueGen with Docker
Instead of installing VueGen locally, you can run it directly from a Docker container with the following command:
```bash
docker run --rm \
-v "$(pwd)/docs/example_data/Earth_microbiome_vuegen_demo_notebook:/home/appuser/Earth_microbiome_vuegen_demo_notebook" \
-v "$(pwd)/output_docker:/home/appuser/quarto_report" \
quay.io/dtu_biosustain_dsp/vuegen:docker --directory /home/appuser/Earth_microbiome_vuegen_demo_notebook --report_type streamlit
```
## Case studies
VueGen’s functionality is demonstrated through two case studies:
**1. Predefined Directory**
This introductory case study uses a predefined directory with plots, dataframes, Markdown, and HTML components. Users can generate reports in different formats and modify the configuration file to customize the report structure.
🔗 [![Open in Colab][colab_badge]][colab_link_intro_demo]
**2. Earth Microbiome Project Data**
This advanced case study demonstrates the application of VueGen in a real-world scenario using data from the [Earth Microbiome Project (EMP)][emp]. The EMP is an initiative to characterize global microbial taxonomic and functional diversity. The notebook process the EMP data, create plots, dataframes, and other components, and organize outputs within a directory to produce reports. Report content and structure can be adapted by modifying the configuration file. Each report consists of sections on exploratory data analysis, metagenomics, and network analysis.
🔗 [![Open in Colab][colab_badge]][colab_link_emp_demo]
> [!NOTE]
> The EMP case study is available online as [HTML][emp-html-demo] and [Streamlit][emp-st-demo] reports.
## Web application deployment
Once a Streamlit report is generated, it can be deployed as a web application to make it accessible online. There are multiple ways to achieve this:
* **Streamlit Community Cloud**: Deploy your report easily using [Streamlit Cloud][st-cloud], as demonstrated in the [EMP VueGen Demo][emp-st-demo]. The process involves moving the necessary scripts, data, and a requirements.txt file into a GitHub repository. Then, the app can be deployed via the Streamlit Cloud interface. The deployment example is available in the `streamlit-report-example` branch.
* **Standalone Executables**: Convert your Streamlit application into a desktop app by packaging it as an executable file for different operating systems. A detailed explanation of this process can be found in this [Streamlit forum post][st-forum-exe].
* **Stlite**: Run Streamlit apps directly in the browser with [stlite][stlite], a WebAssembly port of Streamlit powered by Pyodide, eliminating the need for a server. It also allows packaging apps as standalone desktop executables using stlite desktop.
These options provide flexibility depending on whether the goal is online accessibility, lightweight execution, or local application distribution.
## Contributing
VueGen is an open-source project, and we welcome contributions of all kinds via GitHub issues and pull requests. You can report bugs, suggest improvements, propose new features, or implement changes. Please follow the guidelines in the [CONTRIBUTING](CONTRIBUTING.md) file to ensure that your contribution is easily integrated into the project.
## Credits and acknowledgements
- VueGen was developed by the [Multiomics Network Analytics Group (MoNA)][Mona] at the [Novo Nordisk Foundation Center for Biosustainability (DTU Biosustain)][Biosustain].
- VueGen relies on the work of numerous open-source projects like [Streamlit](streamlit), [Quarto][quarto], and others. A big thank you to their authors for making this possible!
- The vuegen logo was designed based on an image created by [Scriberia][scriberia] for The [Turing Way Community][turingway], which is shared under a CC-BY licence. The original image can be found at [Zenodo][zenodo-turingway].
## Citation
If you use VueGen in your research or publications, please cite it as follows:
**APA:**
Ayala-Ruano, S., Webel, H., & Santos, A. (2025). *VueGen: Automating the generation of scientific reports*. bioRxiv. https://doi.org/10.1101/2025.03.05.641152
**BibTeX:**
```bibtex
@article{Ayala-Ruano2025VueGen,
author = {Ayala-Ruano, Sebastian and Webel, Henry and Santos, Alberto},
title = {VueGen: Automating the generation of scientific reports},
journal = {bioRxiv},
year = {2025},
doi = {10.1101/2025.03.05.641152},
publisher = {Cold Spring Harbor Laboratory},
url = {https://www.biorxiv.org/content/10.1101/2025.03.05.641152},
eprint = {https://www.biorxiv.org/content/10.1101/2025.03.05.641152.full.pdf}
}
```
## Contact and feedback
We appreciate your feedback! If you have any comments, suggestions, or run into issues while using VueGen, feel free to [open an issue][new-issue] in this repository. Your input helps us make VueGen better for everyone.
[streamlit]: https://streamlit.io/
[vuegen-pypi]: https://pypi.org/project/vuegen/
[vuegen-conda]: https://anaconda.org/bioconda/vuegen
[vuegen-docker-quay]: https://quay.io/repository/dtu_biosustain_dsp/vuegen
[vuegen-license]: https://github.com/Multiomics-Analytics-Group/vuegen/blob/main/LICENSE
[vuegen-class-diag-att]: https://raw.githubusercontent.com/Multiomics-Analytics-Group/vuegen/main/docs/images/vuegen_classdiagram_withattmeth.pdf
[vuegen-docs]: https://vuegen.readthedocs.io/
[ci-gh-action]: https://github.com/Multiomics-Analytics-Group/vuegen/actions/workflows/cdci.yml
[ci-docs]: https://github.com/Multiomics-Analytics-Group/vuegen/actions/workflows/docs.yml
[emp-html-demo]: https://multiomics-analytics-group.github.io/vuegen/
[emp-st-demo]: https://earth-microbiome-vuegen-demo.streamlit.app/
[issues]: https://github.com/Multiomics-Analytics-Group/vuegen/issues
[pulls]: https://github.com/Multiomics-Analytics-Group/vuegen/pulls
[vuegen-preprint]: https://doi.org/10.1101/2025.03.05.641152
[quarto]: https://quarto.org/
[quarto-cli-pypi]: https://pypi.org/project/quarto-cli/
[quarto-cli]: https://quarto.org/docs/get-started/
[nfcore]: https://nf-co.re/
[nextflow]: https://www.nextflow.io/
[nf-vuegen]: https://github.com/Multiomics-Analytics-Group/nf-vuegen/
[colab_badge]: https://colab.research.google.com/assets/colab-badge.svg
[colab_link_intro_demo]: https://colab.research.google.com/github/Multiomics-Analytics-Group/vuegen/blob/main/docs/vuegen_basic_case_study.ipynb
[colab_link_emp_demo]: https://colab.research.google.com/github/Multiomics-Analytics-Group/vuegen/blob/main/docs/vuegen_case_study_earth_microbiome.ipynb
[emp]: https://earthmicrobiome.org/
[st-cloud]: https://streamlit.io/cloud
[stlite]: https://github.com/whitphx/stlite
[st-forum-exe]: https://discuss.streamlit.io/t/streamlit-deployment-as-an-executable-file-exe-for-windows-macos-and-android/6812
[Mona]: https://multiomics-analytics-group.github.io/
[Biosustain]: https://www.biosustain.dtu.dk/
[scriberia]: https://www.scriberia.co.uk/
[turingway]: https://github.com/the-turing-way/the-turing-way
[zenodo-turingway]: https://zenodo.org/records/3695300
[new-issue]: https://github.com/Multiomics-Analytics-Group/vuegen/issues/new