https://github.com/natverse/nat
NeuroAnatomy Toolbox: An R package for the (3D) visualisation and analysis of biological image data, especially tracings of single neurons.
https://github.com/natverse/nat
3d connectomics cran image-analysis neuroanatomy neuroanatomy-toolbox neuron neuron-morphology neuroscience r visualisation
Last synced: about 2 months ago
JSON representation
NeuroAnatomy Toolbox: An R package for the (3D) visualisation and analysis of biological image data, especially tracings of single neurons.
- Host: GitHub
- URL: https://github.com/natverse/nat
- Owner: natverse
- Created: 2014-01-02T07:54:01.000Z (over 11 years ago)
- Default Branch: master
- Last Pushed: 2024-02-18T11:01:13.000Z (over 1 year ago)
- Last Synced: 2024-06-17T05:27:14.418Z (about 1 year ago)
- Topics: 3d, connectomics, cran, image-analysis, neuroanatomy, neuroanatomy-toolbox, neuron, neuron-morphology, neuroscience, r, visualisation
- Language: R
- Homepage: https://natverse.org/nat/
- Size: 60.6 MB
- Stars: 61
- Watchers: 17
- Forks: 25
- Open Issues: 61
-
Metadata Files:
- Readme: README.md
- Code of conduct: CODE_OF_CONDUCT.md
Awesome Lists containing this project
- awesome-biological-image-analysis - NeuroAnatomy Toolbox - R package for the (3D) visualisation and analysis of biological image data, especially tracings of single neurons. (Neuroscience)
README
# nat: NeuroAnatomy Toolbox


[](https://natverse.org/)
[](http://dx.doi.org/10.5281/zenodo.10171)
[](https://github.com/natverse/nat/releases/latest)
[](https://cran.r-project.org/package=nat)
[](https://github.com/natverse/nat/actions)
[](https://app.codecov.io/gh/natverse/nat)
[](https://natverse.org//nat/reference/)
[](https://www.r-pkg.org:443/pkg/nat)
An R package for the (3D) visualisation and analysis of biological image data, especially tracings of
single neurons. **nat** is the core package of a wider suite of neuroanatomy
tools introduced at http://natverse.github.io. **nat** (and its [ancestors](https://github.com/jefferis/AnalysisSuite))
have been used in a number of papers from our group including:
[](http://dx.doi.org/10.1016/j.cell.2007.01.040)
[](http://dx.doi.org/10.1016/j.cub.2010.07.045)
[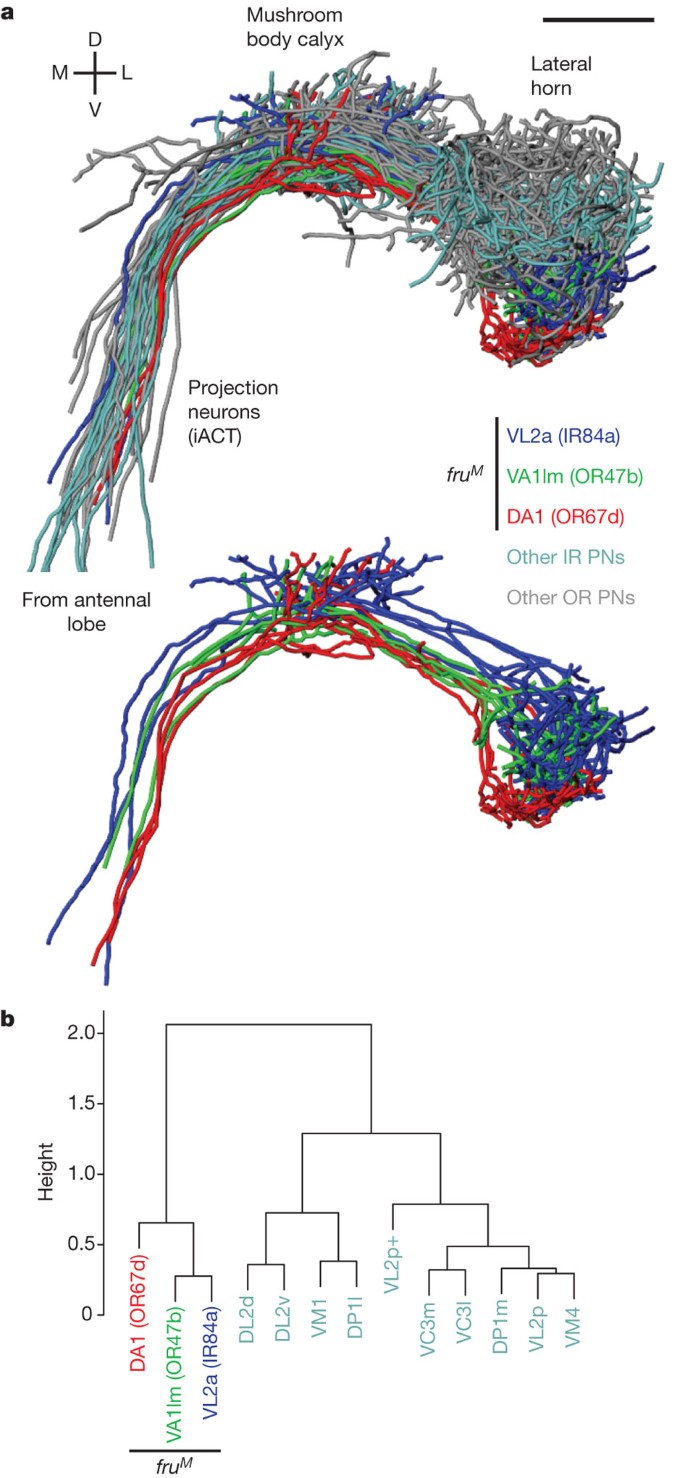 ](http://dx.doi.org/10.1038/nature10428)
](http://dx.doi.org/10.1038/nature10428)
[ ](http://dx.doi.org/10.1016/j.cell.2013.11.025)
](http://dx.doi.org/10.1016/j.cell.2013.11.025)
[](http://dx.doi.org/10.1016/j.neuron.2016.06.012)
## Quick Start
For the impatient ...
```r
# install
install.packages("nat")
# use
library(nat)
# plot some test data (?kcs20 for details)
# Drosophila Kenyon cells processed from raw data at http://flycircuit.tw
head(kcs20)
open3d()
plot3d(kcs20, col=type)
# get help
?nat
```
## Installation
A confirmed stable version of **nat** can be installed from CRAN.
```r
install.packages("nat")
````
However, **nat** remains under quite active development, so if you will be
using **nat** extensively, we generally
recommend installing the latest development version directly from github using
the [devtools](https://cran.r-project.org/package=devtools) package.
```r
# install devtools if required
if (!requireNamespace("devtools")) install.packages("devtools")
# then install nat
devtools::install_github("natverse/nat")
```
## Learn
To get an idea of what **nat** can do:
* Skim through the Articles listed at https://natverse.org//nat/
* Take a look at the [R Markdown](https://rmarkdown.rstudio.com/) reports used
to generate the [figures for our NBLAST paper](http://flybrain.mrc-lmb.cam.ac.uk/si/nblast/www/paper/).
When you're ready to learn more:
* Read the [overview package documentation](https://natverse.org//nat/reference/nat-package.html)
(`?nat` in R)
* Read the [Introduction to neurons](https://natverse.org//nat/articles/neurons-intro.html) article
* Check out the thematically organised [function reference documentation](https://natverse.org//nat/reference/).
* Most help pages include examples.
* Try out sample code:
- [nat.examples](https://github.com/natverse/nat.examples) has detailed
examples for data sets from a range of model organisms and techniques
- [frulhns](https://github.com/jefferis/frulhns) analysis of sexually dimorphic circuits
- [NBLAST figures](https://github.com/jefferislab/NBLAST_figures/)
## Help
If you want some help using **nat**:
* For installation issues, see the [Installation vignette](https://natverse.org//nat/articles/Installation.html)
* Contact [nat-user](https://groups.google.com/forum/#!forum/nat-user) Google group -
we normally respond promptly and you will also be helping future users.
If you think that you have found a bug:
* Install the development version of nat using devtools (see above) and see if
that helps.
* Check the [github issues](https://github.com/natverse/nat/issues?q=is%3Aissue) and
- [file a bug report](https://github.com/natverse/nat/issues/new) if this seems to be a new problem
- comment on an existing bug report
Thanks for your interest in **nat**!