https://github.com/neuml/paperai
📄 🤖 Semantic search and workflows for medical/scientific papers
https://github.com/neuml/paperai
ai artificial-intelligence document-search machine-learning medical nlp python scientific-papers search txtai
Last synced: 4 months ago
JSON representation
📄 🤖 Semantic search and workflows for medical/scientific papers
- Host: GitHub
- URL: https://github.com/neuml/paperai
- Owner: neuml
- License: apache-2.0
- Created: 2020-07-21T18:33:30.000Z (over 5 years ago)
- Default Branch: master
- Last Pushed: 2025-04-21T17:36:05.000Z (10 months ago)
- Last Synced: 2025-04-21T18:30:50.068Z (10 months ago)
- Topics: ai, artificial-intelligence, document-search, machine-learning, medical, nlp, python, scientific-papers, search, txtai
- Language: Python
- Homepage:
- Size: 1.73 MB
- Stars: 1,394
- Watchers: 24
- Forks: 108
- Open Issues: 0
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
README

AI for medical and scientific papers
-------------------------------------------------------------------------------------------------------------------------------------------------------
`paperai` is an AI application for medical and scientific papers.

⚡ Supercharge research tasks with AI-driven report generation. A `paperai` application goes through repositories of articles and generates bulk answers to questions backed by Large Language Model (LLM) prompts and Retrieval Augmented Generation (RAG) pipelines.
A `paperai` configuration file enables bulk LLM inference operations in a performant manner. Think of it like kicking off hundreds of ChatGPT prompts over your data.

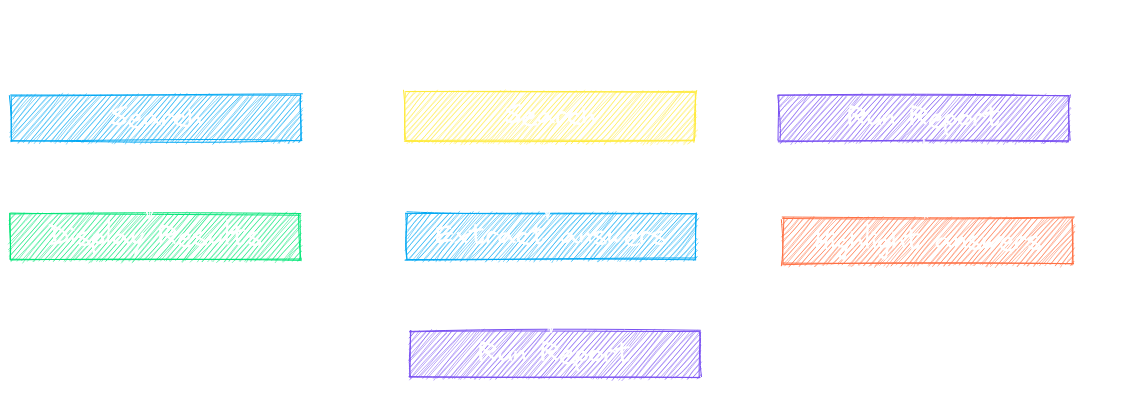
`paperai` can generate reports in Markdown, CSV and annotate answers directly on PDFs (when available).
## Installation
The easiest way to install is via pip and PyPI
```
pip install paperai
```
Python 3.10+ is supported. Using a Python [virtual environment](https://docs.python.org/3/library/venv.html) is recommended.
`paperai` can also be installed directly from GitHub to access the latest, unreleased features.
```
pip install git+https://github.com/neuml/paperai
```
See [this link](https://neuml.github.io/txtai/install/#environment-specific-prerequisites) to help resolve environment-specific install issues.
### Docker
Run the steps below to build a docker image with `paperai` and all dependencies.
```
wget https://raw.githubusercontent.com/neuml/paperai/master/docker/Dockerfile
docker build -t paperai .
docker run --name paperai --rm -it paperai
```
paperetl can be added in to have a single image to index and query content. Follow the instructions to build a [paperetl docker image](https://github.com/neuml/paperetl#docker) and then run the following.
```
docker build -t paperai --build-arg BASE_IMAGE=paperetl --build-arg START=/scripts/start.sh .
docker run --name paperai --rm -it paperai
```
## Examples
The following notebooks and applications demonstrate the capabilities provided by `paperai`.
### Notebooks
| Notebook | Description | |
|:----------|:-------------|------:|
| [Introducing paperai](https://github.com/neuml/paperai/blob/master/examples/01_Introducing_paperai.ipynb) | Overview of the functionality provided by paperai | [](https://colab.research.google.com/github/neuml/paperai/blob/master/examples/01_Introducing_paperai.ipynb) |
| [Medical Research Project](https://github.com/neuml/paperai/blob/master/examples/02_Medical_Research_Project.ipynb) | Research young onset colon cancer | [](https://colab.research.google.com/github/neuml/paperai/blob/master/examples/02_Medical_Research_Project.ipynb) |
### Applications
| Application | Description |
|:----------|:-------------|
| [Search](https://github.com/neuml/paperai/blob/master/examples/search.py) | Search a `paperai` index. Set query parameters, execute searches and display results. |
## Building a model
`paperai` indexes databases previously built with [paperetl](https://github.com/neuml/paperetl). The following shows how to create a new `paperai` index.
1. (Optional) Create an index.yml file
`paperai` uses the default txtai embeddings configuration when not specified. Alternatively, an index.yml file can be specified that takes all the same options as a txtai embeddings instance. See the [txtai documentation](https://neuml.github.io/txtai/embeddings/configuration) for more on the possible options. A simple example is shown below.
```
path: sentence-transformers/all-MiniLM-L6-v2
content: True
```
2. Build embeddings index
```
python -m paperai.index
```
The paperai.index process requires an input data path and optionally takes index configuration. This configuration can either be a vector model path or an index.yml configuration file.
## Running queries
The fastest way to run queries is to start a `paperai` shell
```
paperai
```
A prompt will come up. Queries can be typed directly into the console.
## Report schema
The following steps through an example `paperai` report configuration file and describes each section.
```yaml
name: ColonCancer
options:
llm: Intelligent-Internet/II-Medical-8B-1706-GGUF/II-Medical-8B-1706.Q4_K_M.gguf
system: You are a medical literature document parser. You extract fields from data.
template: |
Quickly extract the following field using the provided rules and context.
Rules:
- Keep it simple, don't overthink it
- ONLY extract the data
- NEVER explain why the field is extracted
- NEVER restate the field name only give the field value
- Say no data if the field can't be found within the context
Field:
{question}
Context:
{context}
context: 5
params:
maxlength: 4096
stripthink: True
Research:
query: colon cancer young adults
columns:
- name: Date
- name: Study
- name: Study Link
- name: Journal
- {name: Sample Size, query: number of patients, question: Sample Size}
- {name: Objective, query: objective, question: Study Objective}
- {name: Causes, query: possible causes, question: List of possible causes}
- {name: Detection, query: diagnosis, question: List of ways to diagnose}
```
### Configuration
The following shows the top level configuration options.
| Field | Description |
|:------------ |:-------------|
| name | Report name |
| options | RAG pipeline options - set the LLM, prompt templates, max length and more|
| report | Each unique top level parameter sets the report name. In the example above, it's called `Research` |
| query | Vector query that identifies the top n documents |
| columns | List of columns |
### Standard columns
Standard columns use the article data store metadata to simply copy fields into a report. Set the column `name` to one of the values below.
| Field | Description |
|:------------ |:-------------|
| Id | Article unique identifier |
| Date | Article publication date |
| Study | Title of the article |
| Study Link | HTTP link to the study |
| Journal | Publication name |
| Source | Data source name |
| Entry | Article entry date |
| Matches | Sections that caused this article to match the report query |
### Generated columns
The most novel feature of `paperai` is being able to generate dynamic columns driven by a RAG pipeline. Each field takes the following parameters.
| Parameter | Description |
|:------------ |:-------------|
| name | Column name |
| query | search/similarity query |
| question | llm question parameter |
For each matching article, the `query` sorts each section by relevance to that query. This can be a vector query, keyword query or hybrid query. This is controlled by the embeddings index configuration. The `question` is plugged into the RAG pipeline template along with the top n matching context elements from the query. The generated column is stored as `name` in the report output.
## Building a report file
Reports can generate output in multiple formats. An example report call:
```
python -m paperai.report crc.yml 10 csv
```
In the example above, a file named Research.csv will be created with the top 10 most relevant articles.
The following report formats are supported:
- Markdown (Default) - Renders a Markdown report. Columns and answers are extracted from articles with the results stored in a Markdown file.
- CSV - Renders a CSV report. Columns and answers are extracted from articles with the results stored in a CSV file.
- Annotation - Columns and answers are extracted from articles with the results annotated over the original PDF files. Requires passing in a path with the original PDF files.
See the [examples](https://github.com/neuml/paperai/tree/master/examples) directory for report examples. Additional historical report configuration files can be found [here](https://github.com/neuml/cord19q/tree/master/tasks).
## Tech Overview
`paperai` is a combination of a [txtai](https://github.com/neuml/txtai) embeddings index, a SQLite database with the articles and an LLM. These components are joined together in a [txtai RAG pipeline](https://neuml.github.io/txtai/pipeline/text/rag/).
Each article is parsed into sections and stored in a data store along with the article metadata. Embeddings are built over the full corpus. The LLM analyzes context-limited requests and generates outputs.
Multiple entry points exist to interact with the model.
- paperai.report - Builds a report for a series of queries. For each query, the top scoring articles are shown along with matches from those articles. There is also a highlights section showing the most relevant results.
- paperai.query - Runs a single query from the terminal
- paperai.shell - Allows running multiple queries from the terminal
## Recognition
`paperai` and/or NeuML has been recognized in the following articles.
- [Machine-Learning Experts Delve Into 47,000 Papers on Coronavirus Family](https://www.wsj.com/articles/machine-learning-experts-delve-into-47-000-papers-on-coronavirus-family-11586338201)
- [Data scientists assist medical researchers in the fight against COVID-19](https://cloud.google.com/blog/products/ai-machine-learning/how-kaggle-data-scientists-help-with-coronavirus)
- [CORD-19 Kaggle Challenge Awards](https://www.kaggle.com/allen-institute-for-ai/CORD-19-research-challenge/discussion/161447)




