https://github.com/neuronets/nobrainer
A framework for developing neural network models for 3D image processing.
https://github.com/neuronets/nobrainer
machine-learning medical-imaging neuroimaging semantic-segmentation tensorflow
Last synced: 2 months ago
JSON representation
A framework for developing neural network models for 3D image processing.
- Host: GitHub
- URL: https://github.com/neuronets/nobrainer
- Owner: neuronets
- License: other
- Created: 2017-06-16T23:38:47.000Z (about 8 years ago)
- Default Branch: master
- Last Pushed: 2025-03-31T18:05:50.000Z (4 months ago)
- Last Synced: 2025-04-08T13:11:12.679Z (3 months ago)
- Topics: machine-learning, medical-imaging, neuroimaging, semantic-segmentation, tensorflow
- Language: Python
- Homepage:
- Size: 2.42 MB
- Stars: 160
- Watchers: 9
- Forks: 43
- Open Issues: 61
-
Metadata Files:
- Readme: README.md
- Changelog: CHANGELOG.md
- License: LICENSE
- Citation: CITATION
Awesome Lists containing this project
README
# Nobrainer

_Nobrainer_ is a deep learning framework for 3D image processing. It implements
several 3D convolutional models from recent literature, methods for loading and
augmenting volumetric data that can be used with any TensorFlow or Keras model,
losses and metrics for 3D data, and simple utilities for model training, evaluation,
prediction, and transfer learning.
_Nobrainer_ also provides pre-trained models for brain extraction, brain segmentation,
brain generation and other tasks. Please see the [_Trained_ models](https://github.com/neuronets/trained-models)
repository for more information.
The _Nobrainer_ project is supported by NIH RF1MH121885 and is distributed under
the Apache 2.0 license. It was started under the support of NIH R01 EB020470.
## Table of contents
- [Implementations](#implementations)
- [Models](#models)
- [Dropout and additional layers](#dropout-and-regularization-layers)
- [Losses](#losses)
- [Metrics](#metrics)
- [Augmentation methods](#augmentation-methods)
- [Guide Jupyter Notebooks](#guide-jupyter-notebooks-)
- [Installation](#installation)
- [Container](#container)
- [GPU support](#gpu-support)
- [CPU only](#cpu-only)
- [pip](#pip)
- [Using pre-trained networks](#using-pre-trained-networks)
- [Predicting a brainmask for a T1-weighted brain scan](#predicting-a-brainmask-for-a-t1-weighted-brain-scan)
- [Generating a synthetic T1-weighted brain scan](#generating-a-synthetic-t1-weighted-brain-scan)
- [Transfer learning](#transfer-learning)
- [Data augmentation](#data-augmentation)
- [Random rigid transformation](#random-rigid-transformation)
- [Package layout](#package-layout)
- [Questions or issues](#questions-or-issues)
## Implementations
### Models
| Model | Type | Application |
|:-----------|:-------------:|:-------------:|
|[**Highresnet**](https://github.com/neuronets/nobrainer/blob/master/nobrainer/models/highresnet.py) [(source)](https://arxiv.org/abs/1707.01992)| supervised | segmentation/classification |
|[**Unet**](https://github.com/neuronets/nobrainer/blob/master/nobrainer/models/unet.py) [(source)](https://arxiv.org/abs/1606.06650)| supervised | segmentation/classification |
|[**Vnet**](https://github.com/neuronets/nobrainer/blob/master/nobrainer/models/vnet.py) [(source)](https://arxiv.org/pdf/1606.04797.pdf)| supervised | segmentation/classification |
|[**Meshnet**](https://github.com/neuronets/nobrainer/blob/master/nobrainer/models/meshnet.py) [(source)](https://arxiv.org/abs/1612.00940)| supervised | segmentation/clssification |
|[**Bayesian Meshnet**](https://github.com/neuronets/nobrainer/blob/master/nobrainer/models/bayesian-meshnet.py) [(source)](https://www.frontiersin.org/articles/10.3389/fninf.2019.00067/full)| bayesian supervised | segmentation/classification |
|[**Bayesian Vnet**](https://github.com/neuronets/nobrainer/blob/master/nobrainer/models/bayesian_vnet.py) | bayesian supervised | segmentation/classification |
|[**Semi_Bayesian Vnet**](https://github.com/neuronets/nobrainer/blob/master/nobrainer/models/bayesian_vnet.py) | semi-bayesian supervised | segmentation/classification |
|[**DCGAN**](https://github.com/neuronets/nobrainer/blob/master/nobrainer/models/dcgan.py) | self supervised | generative model |
|[**Progressive GAN**](https://github.com/neuronets/nobrainer/blob/master/nobrainer/models/progressivegan.py) | self supervised | generative model |
|[**3D Autoencoder**](https://github.com/neuronets/nobrainer/blob/master/nobrainer/models/autoencoder.py) | self supervised | knowledge representation/dimensionality reduction |
|[**3D Progressive Autoencoder**](https://github.com/neuronets/nobrainer/blob/master/nobrainer/models/progressiveae.py) | self supervised | knowledge representation/dimensionality reduction |
|[**3D SimSiam**](https://github.com/neuronets/nobrainer/blob/master/nobrainer/models/brainsiam.py) [(source)](https://arxiv.org/abs/2011.10566)| self supervised | Siamese Representation Learning |
### Dropout and regularization layers
[Bernouli dropout layer](https://github.com/neuronets/nobrainer/blob/80d8a47a7f2bf4fe335bdf194c0be19044223629/nobrainer/layers/dropout.py#L15), [Concrete dropout layer](https://github.com/neuronets/nobrainer/blob/80d8a47a7f2bf4fe335bdf194c0be19044223629/nobrainer/layers/dropout.py#L71), [Gaussian dropout](https://github.com/neuronets/nobrainer/blob/80d8a47a7f2bf4fe335bdf194c0be19044223629/nobrainer/layers/dropout.py#L204), [Group normalization layer](), [Costom padding layer]()
### Losses
[Dice](), [Jaccard](), [Tversky](), [ELBO](), [Wasserstien](), [Gradient Penalty]()
### Metrics
[Dice](), [Generalized Dice](), [Jaccard](), [Hamming](), [Tversky]()
### Augmentation methods
#### Spatial Transforms
[Center crop](), [Spatial Constant Padding](), [Random Crop](), [Resize](), [Random flip (left and right)]()
#### Intensity Transforms
[Add gaussian noise](), [Min-Max intensity scaling](), [Costom intensity scaling](), [Intensity masking](), [Contrast adjustment]()
#### Affine Transform
Afifine transformation including rotation, translation, reflection.
## Guide Jupyter Notebooks [](https://colab.research.google.com/github/neuronets/nobrainer-book/blob/master/)
Please refer to the Jupyter notebooks in the [guide](https://github.com/neuronets/nobrainer-book/blob/master/docs/nobrainer-guides/notebooks/) directory to get
started with _Nobrainer_. [Try them out](https://colab.research.google.com/github/neuronets/nobrainer-book/blob/master/) in Google Colaboratory!
## Installation
### Container
We recommend using the official _Nobrainer_ Docker container, which includes all
of the dependencies necessary to use the framework. Please see the available images
on [DockerHub](https://hub.docker.com/r/neuronets/nobrainer)
#### GPU support
The _Nobrainer_ containers with GPU support use the Tensorflow jupyter GPU containers. Please
check the containers for the version of CUDA installed. Nvidia drivers are not included in
the container.
```
$ docker pull neuronets/nobrainer:latest-gpu
$ singularity pull docker://neuronets/nobrainer:latest-gpu
```
#### CPU only
This container can be used on all systems that have Docker or Singularity and
does not require special hardware. This container, however, should not be used
for model training (it will be very slow).
```
$ docker pull neuronets/nobrainer:latest-cpu
$ singularity pull docker://neuronets/nobrainer:latest-cpu
```
### pip
_Nobrainer_ can also be installed with pip.
```
$ pip install nobrainer
```
## Using pre-trained networks
Pre-trained networks are available in the [_Trained_ models](https://github.com/neuronets/trained-models)
repository. Prediction can be done on the command-line with `nobrainer predict`
or in Python. Similarly, generation can be done on the command-line with
`nobrainer generate` or in Python.
### Predicting a brainmask for a T1-weighted brain scan
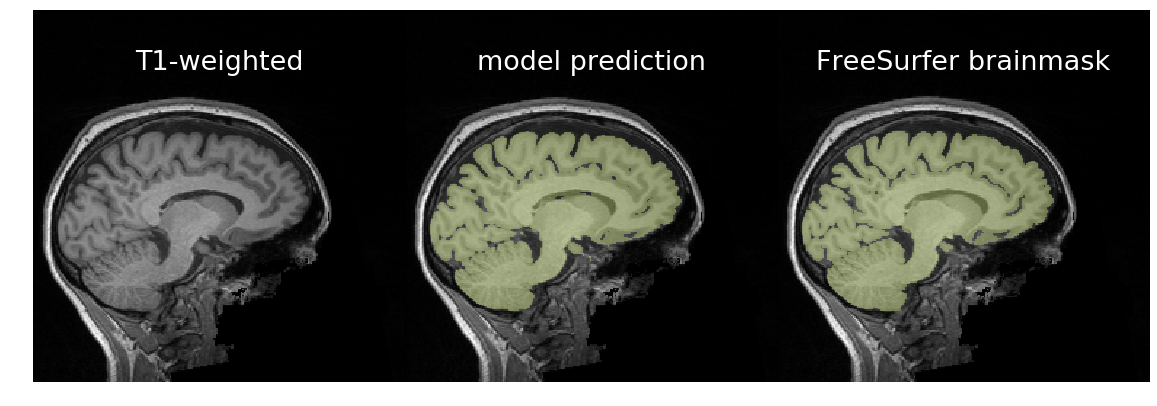
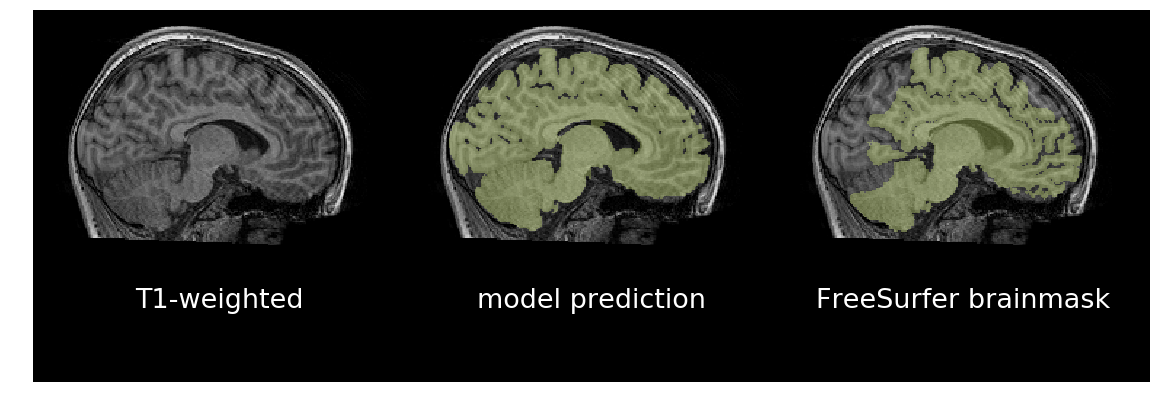
__Figure__: In the first column are T1-weighted brain scans, in the middle
are a trained model's predictions, and on the right are binarized FreeSurfer
segmentations. Despite being trained on binarized FreeSurfer segmentations,
the model outperforms FreeSurfer in the bottom scan, which exhibits motion
distortion. It took about three seconds for the model to predict each brainmask
using an NVIDIA GTX 1080Ti. It takes about 70 seconds on a recent CPU.
In the following examples, we will use a 3D U-Net trained for brain extraction and
documented in [_Trained_ models](https://github.com/neuronets/trained-models#brain-extraction).
In the base case, we run the T1w scan through the model for prediction.
```bash
# Get sample T1w scan.
wget -nc https://dl.dropbox.com/s/g1vn5p3grifro4d/T1w.nii.gz
docker run --rm -v $PWD:/data neuronets/nobrainer \
predict \
--model=/models/neuronets/brainy/0.1.0/brain-extraction-unet-128iso-model.h5 \
--verbose \
/data/T1w.nii.gz \
/data/brainmask.nii.gz
```
For binary segmentation where we expect one predicted region, as is the case with
brain extraction, we can reduce false positives by removing all predictions not
connected to the largest contiguous label.
```bash
# Get sample T1w scan.
wget -nc https://dl.dropbox.com/s/g1vn5p3grifro4d/T1w.nii.gz
docker run --rm -v $PWD:/data neuronets/nobrainer \
predict \
--model=/models/neuronets/brainy/0.1.0/brain-extraction-unet-128iso-model.h5 \
--largest-label \
--verbose \
/data/T1w.nii.gz \
/data/brainmask-largestlabel.nii.gz
```
Because the network was trained on randomly rotated data, it should be agnostic
to orientation. Therefore, we can rotate the volume, predict on it, undo the
rotation in the prediction, and average the prediction with that from the original
volume. This can lead to a better overall prediction but will at least double the
processing time. To enable this, use the flag `--rotate-and-predict` in
`nobrainer predict`.
```bash
# Get sample T1w scan.
wget -nc https://dl.dropbox.com/s/g1vn5p3grifro4d/T1w.nii.gz
docker run --rm -v $PWD:/data neuronets/nobrainer \
predict \
--model=/models/neuronets/brainy/0.1.0/brain-extraction-unet-128iso-model.h5 \
--rotate-and-predict \
--verbose \
/data/T1w.nii.gz \
/data/brainmask-withrotation.nii.gz
```
Combining the above, we can usually achieve the best brain extraction by using
`--rotate-and-predict` in conjunction with `--largest-label`.
```bash
# Get sample T1w scan.
wget -nc https://dl.dropbox.com/s/g1vn5p3grifro4d/T1w.nii.gz
docker run --rm -v $PWD:/data neuronets/nobrainer \
predict \
--model=/models/neuronets/brainy/0.1.0/brain-extraction-unet-128iso-model.h5 \
--largest-label \
--rotate-and-predict \
--verbose \
/data/T1w.nii.gz \
/data/brainmask-maybebest.nii.gz
```
### Generating a synthetic T1-weighted brain scan
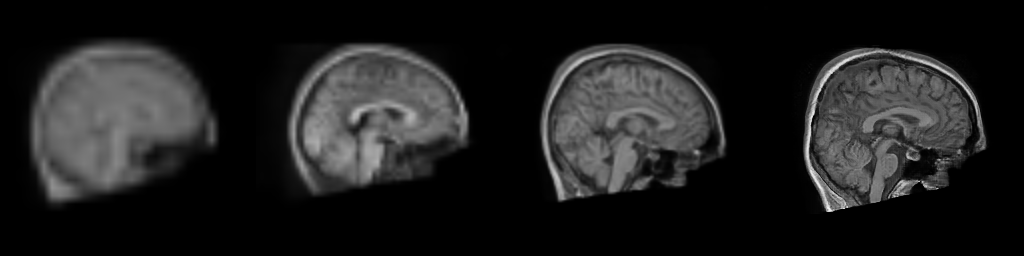
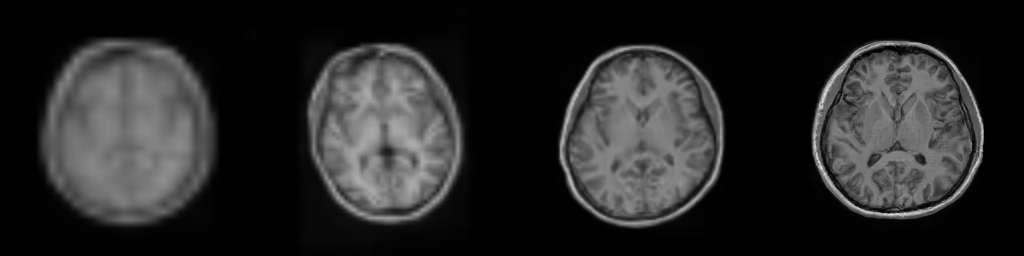
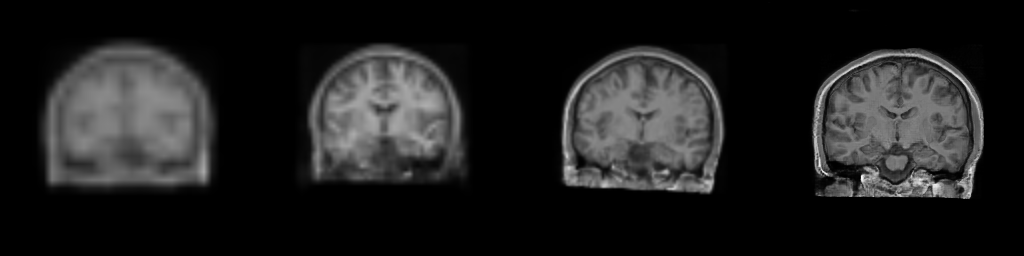
__Figure__: Progressive generation of T1-weighted brain MR scan starting
from a resolution of 32 to 256 (Left to Right: 323, 643,
1283, 2563). The brain scans are generated using the same
latents in all resolutions. It took about 6 milliseconds for the model to generate
the 2563 brainscan using an NVIDIA TESLA V-100.
In the following examples, we will use a Progressive Generative Adversarial Network
trained for brain image generation and documented in
[_Trained_ models](https://github.com/neuronets/trained-models#brain-extraction).
In the base case, we generate a T1w scan through the model for a given resolution.
We need to pass the directory containing the models `(tf.SavedModel)` created
while training the networks.
```bash
docker run --rm -v $PWD:/data neuronets/nobrainer \
generate \
--model=/models/neuronets/braingen/0.1.0 \
--output-shape=128 128 128 \
/data/generated.nii.gz
```
We can also generate multiple resolutions of the brain image using the same
latents to visualize the progression
```bash
# Get sample T1w scan.
docker run --rm -v $PWD:/data neuronets/nobrainer \
generate \
--model=/models/neuronets/braingen/0.1.0 \
--multi-resolution \
/data/generated.nii.gz
```
In the above example, the multi resolution images will be saved as
`generated_res_{resolution}.nii.gz`
### Transfer learning
The pre-trained models can be used for transfer learning. To avoid forgetting
important information in the pre-trained model, you can apply regularization to
the kernel weights and also use a low learning rate. For more information, please
see the _Nobrainer_ guide notebook on transfer learning.
As an example of transfer learning, [@kaczmarj](https://github.com/kaczmarj)
re-trained a brain extraction model to label meningiomas in 3D T1-weighted,
contrast-enhanced MR scans. The original model is publicly available and was
trained on 10,000 T1-weighted MR brain scans from healthy participants. These
were all research scans (i.e., non-clinical) and did not include any contrast
agents. The meningioma dataset, on the other hand, was composed of relatively
few scans, all of which were clinical and used gadolinium as a contrast agent.
You can observe the differences in contrast below.


Despite the differences between the two datasets, transfer learning led to a much
better model than training from randomly-initialized weights. As evidence, please
see below violin plots of Dice coefficients on a validation set. In the left plot
are Dice coefficients of predictions obtained with the model trained from
randomly-initialized weights, and on the right are Dice coefficients of predictions
obtained with the transfer-learned model. In general, Dice coefficients are higher
on the right, and the variance of Dice scores is lower. Overall, the model on the
right is more accurate and more robust than the one on the left.


## Package layout
- `nobrainer.io`: input/output methods
- `nobrainer.layers`: custom layers, which conform to the Keras API
- `nobrainer.losses`: loss functions for volumetric segmentation
- `nobrainer.metrics`: metrics for volumetric segmentation
- `nobrainer.models`: pre-defined Keras models
- `nobrainer.training`: training utilities (supports training on single and multiple GPUs)
- `nobrainer.transform`: random rigid transformations for data augmentation
- `nobrainer.volume`: `tf.data.Dataset` creation and data augmentation utilities
-
## Citation
If you use this package, please [cite](https://github.com/neuronets/nobrainer/blob/master/CITATION) it.
## Questions or issues
If you have questions about _Nobrainer_ or encounter any issues using the framework,
please [submit a GitHub issue](https://github.com/neuronets/helpdesk/issues/new/choose).