https://github.com/nlmixr2/nonmem2rx
nonmem2rx
https://github.com/nlmixr2/nonmem2rx
nlmixr2 nonmem pharmacometrics rxode2
Last synced: about 2 months ago
JSON representation
nonmem2rx
- Host: GitHub
- URL: https://github.com/nlmixr2/nonmem2rx
- Owner: nlmixr2
- Created: 2023-01-25T19:11:05.000Z (over 2 years ago)
- Default Branch: main
- Last Pushed: 2024-05-28T23:18:11.000Z (over 1 year ago)
- Last Synced: 2024-05-29T01:48:35.771Z (over 1 year ago)
- Topics: nlmixr2, nonmem, pharmacometrics, rxode2
- Language: ReScript
- Homepage: https://nlmixr2.github.io/nonmem2rx/
- Size: 81.8 MB
- Stars: 9
- Watchers: 6
- Forks: 0
- Open Issues: 8
-
Metadata Files:
- Readme: README.Rmd
Awesome Lists containing this project
README
---
output: github_document
---
```{r, include = FALSE}
knitr::opts_chunk$set(
collapse = TRUE,
comment = "#>",
fig.path = "man/figures/README-",
out.width = "100%"
)
```
# nonmem2rx
[](https://github.com/nlmixr2/nonmem2rx/actions/workflows/R-CMD-check.yaml)
[](https://app.codecov.io/gh/nlmixr2/nonmem2rx?branch=main)
[](https://cran.r-project.org/package=nonmem2rx)
[](https://cran.r-project.org/package=nonmem2rx)
[](https://cran.r-project.org/package=nonmem2rx)
[](https://www.codefactor.io/repository/github/nlmixr2/nonmem2rx)

The goal of nonmem2rx is to convert a NONMEM control stream to a
`rxode2` (or even a `nlmixr2` fit) for easy clinical trial simulation
in R.
## Installation
You can install the development version of nonmem2rx from
[GitHub](https://github.com/) with the r-universe:
``` r
install.packages('nonmem2rx', repos = c('https://nlmixr2.r-universe.dev', 'https://cloud.r-project.org'))
```
You can also get the CRAN version by:
``` r
install.packages('nonmem2rx')
```
## What you can do with `nonmem2rx`/`babelmixr2`
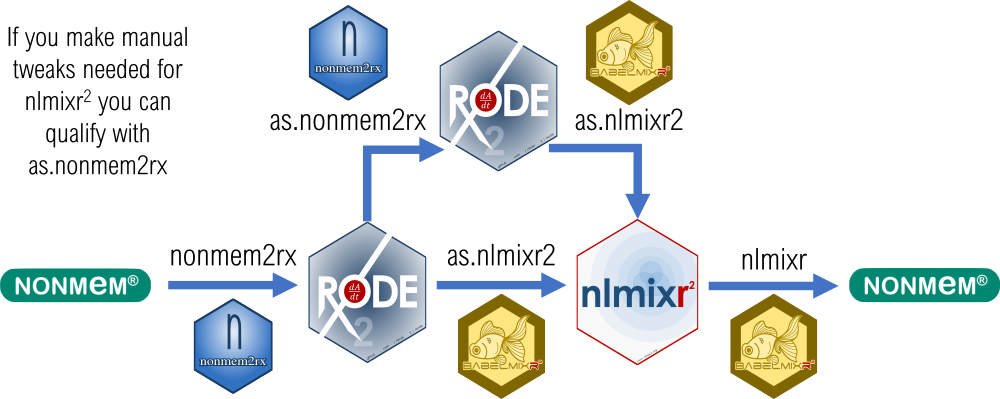
You can do many useful tasks directly converting between nlmixr2 and NONMEM models; you can:
- [Convert a NONMEM model to a rxode2 model](https://nlmixr2.github.io/nonmem2rx/articles/import-nonmem.html)
- [Do development in nlmixr2 and then run NONMEM from a nlmixr2 model](https://nlmixr2.github.io/babelmixr2/articles/running-nonmem.html) for reviewers who want to know about NONMEM results.
- In both conversions, [automatically make sure the model is translated correctly ](https://nlmixr2.github.io/nonmem2rx/articles/rxode2-validate.html) (for [babelmixr2](https://nlmixr2.github.io/babelmixr2/articles/running-nonmem.html#optional-step-2-recover-a-failed-nonmem-run))
Then with nlmixr2 fit models and nonmem2rx models coming from both conversions, you can:
- [Perform simulations of new
dosing](https://nlmixr2.github.io/nonmem2rx/articles/simulate-new-dosing.html) from the NONMEM model or even [simulate using the uncertainty in your model to simulate new scenarios](https://nlmixr2.github.io/nonmem2rx/articles/simulate-uncertainty.html)
- [Modify the model to calculate derived parameters](https://nlmixr2.github.io/nonmem2rx/articles/simulate-extra-items.html) (like AUC). These parameters slow down NONMEM’s optimization, but can help in your simulation scenario.
- [Simulating with Covariates/Input PK
parameters](https://nlmixr2.github.io/nonmem2rx/articles/simulate-new-dosing-with-covs.html). This example shows approaches to resample from the input dataset for covariate selection.
With nonmem2rx and babelmixr2, convert the imported rxode2 model to a nlmixr2 object, allowing:
- [Generation of Word and PowerPoint plots with
nlmixr2rpt](https://nlmixr2.github.io/nonmem2rx/articles/create-office.html)
- [Easy VPC
creation](https://nlmixr2.github.io/nonmem2rx/articles/create-vpc.html)
(with `vpcPlot()`)
- [Easy Individual plots with extra solved
points](https://nlmixr2.github.io/nonmem2rx/articles/create-augPred.html). This
will show the curvature of individual and population fits for
sparse data-sets (with `augPred()`)
You can even use this conversion to help debug your NONMEM model (or
even try it in nlmixr2 instead)
- [Understand how to simplify the NONMEM model to avoid rounding errors](https://nlmixr2.github.io/nonmem2rx/articles/read-rounding.html)
- [Run nlmixr2’s covariance step when NONMEMs covariance step has failed](https://nlmixr2.github.io/nonmem2rx/articles/read-rounding.html#step-5-get-the-covariance-of-the-model) (in the linked example, there was no covariance step because rounding errors)
## Simple example
Once `nonmem2rx` has been loaded, you simply type the location of the
nonmem control stream for the parser to start. For example:
```{r example}
library(nonmem2rx)
# First we need the location of the nonmem control stream Since we are
# running an example, we will use one of the built-in examples in
# `nonmem2rx`
ctlFile <- system.file("mods/cpt/runODE032.ctl", package="nonmem2rx")
# You can use a control stream or other file. With the development
# version of `babelmixr2`, you can simply point to the listing file
mod <- nonmem2rx(ctlFile, lst=".res", save=FALSE)
mod
```
You can see this automatically validates NONMEM and rxode2 outputs for
a couple of metrics.
# External projects that contributed to the tool's validation
The `nonmem2rx` tool was validated against:
- The `PsN` library test suite of NONMEM listings
(https://github.com/UUPharmacometrics/PsN/tree/master/test)
- The ddmore model scrapings
(https://github.com/dpastoor/ddmore_scraping).
- Models from NONMEM design tutorial Bauer 2021 https://doi.org/10.1002/psp4.12713
- Models from NONMEM tutorial 1 (Bauer 2019) https://doi.org/10.1002/psp4.12404
- Models from NONMEM tutorial 2 (Bauer 2019) https://doi.org/10.1002/psp4.12422
Due to the sheer size of the zipped models for these nonmem control
stream sources, these are excluded to keep the binary below 3 mgs
(CRAN requirement).
However, I would like to acknowledge all who helped in these projects.
With these projects the NONMEM conversion to rxode2 has been made much
more robust.
Still, while the tests are not/will not be in the CRAN binaries, you
can test them yourself by:
1. Downloading this repository
2. Running the tests `devtools::test()`