https://github.com/osscar-org/jupyterlab-mol-visualizer
A JupyterLab launcher extension to view the molecular orbitals.
https://github.com/osscar-org/jupyterlab-mol-visualizer
jupyterlab jupyterlab-extension molecular-ortibal
Last synced: 6 months ago
JSON representation
A JupyterLab launcher extension to view the molecular orbitals.
- Host: GitHub
- URL: https://github.com/osscar-org/jupyterlab-mol-visualizer
- Owner: osscar-org
- License: other
- Created: 2021-08-23T20:30:55.000Z (about 4 years ago)
- Default Branch: main
- Last Pushed: 2024-09-12T06:51:31.000Z (about 1 year ago)
- Last Synced: 2025-03-26T23:21:58.337Z (7 months ago)
- Topics: jupyterlab, jupyterlab-extension, molecular-ortibal
- Language: TypeScript
- Homepage:
- Size: 9.66 MB
- Stars: 16
- Watchers: 1
- Forks: 2
- Open Issues: 1
-
Metadata Files:
- Readme: README.md
- Changelog: CHANGELOG.md
- License: LICENSE
Awesome Lists containing this project
README
# `jupyterlab-mol-visualizer`: A JupyterLab Extension to Visualize Molecular Orbitals
[](https://github.com/osscar-org/jupyterlab-mol-visualizer/actions/workflows/build.yml)
A JupyterLab launcher extension to view the molecular orbitals (MOs).
[NGL](https://github.com/nglviewer/ngl) JavaScript package was employed to visulizer the MOs.
The icon of the extension is from [SVG Repo](https://www.svgrepo.com/svg/231591/molecule).
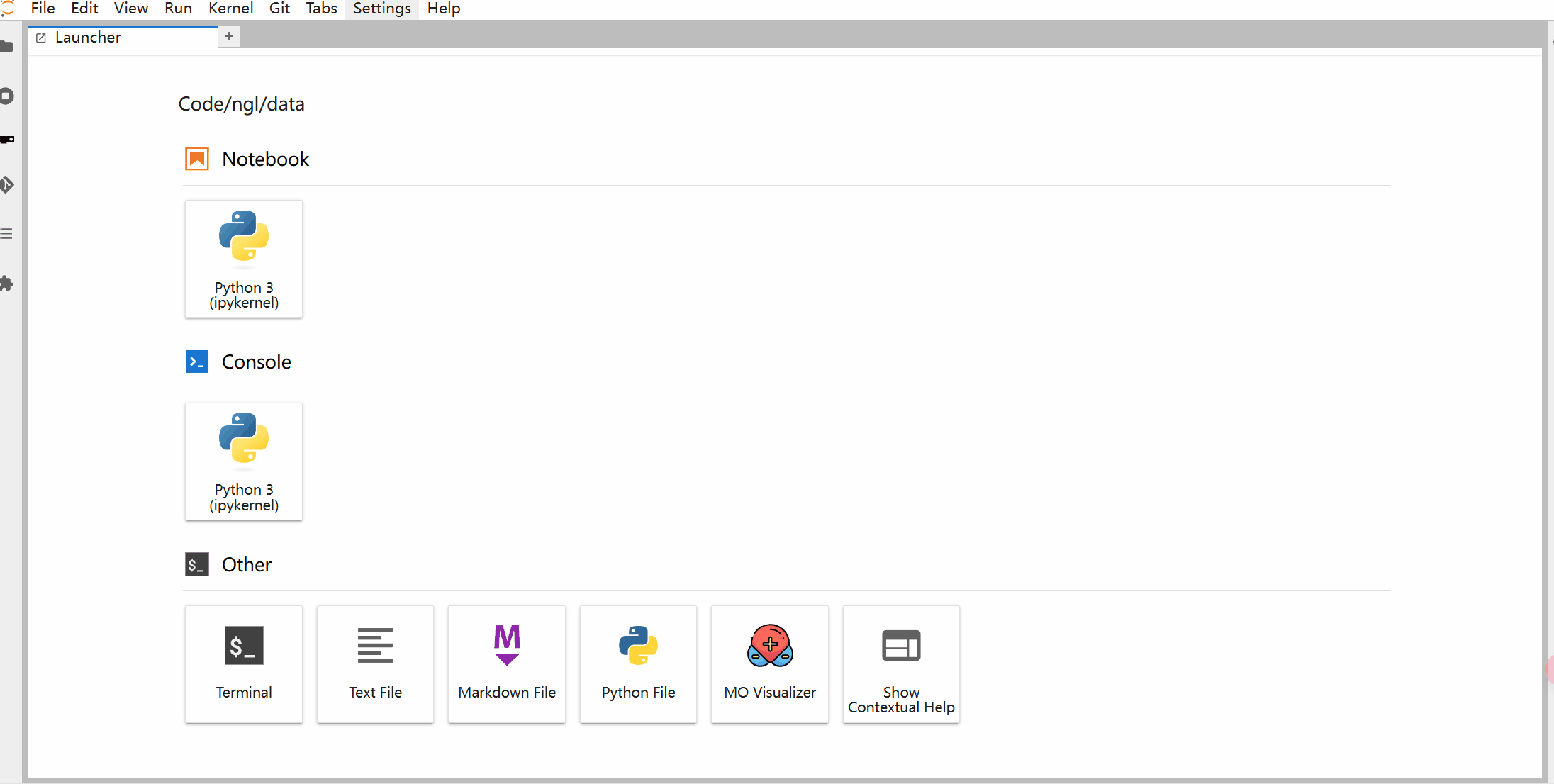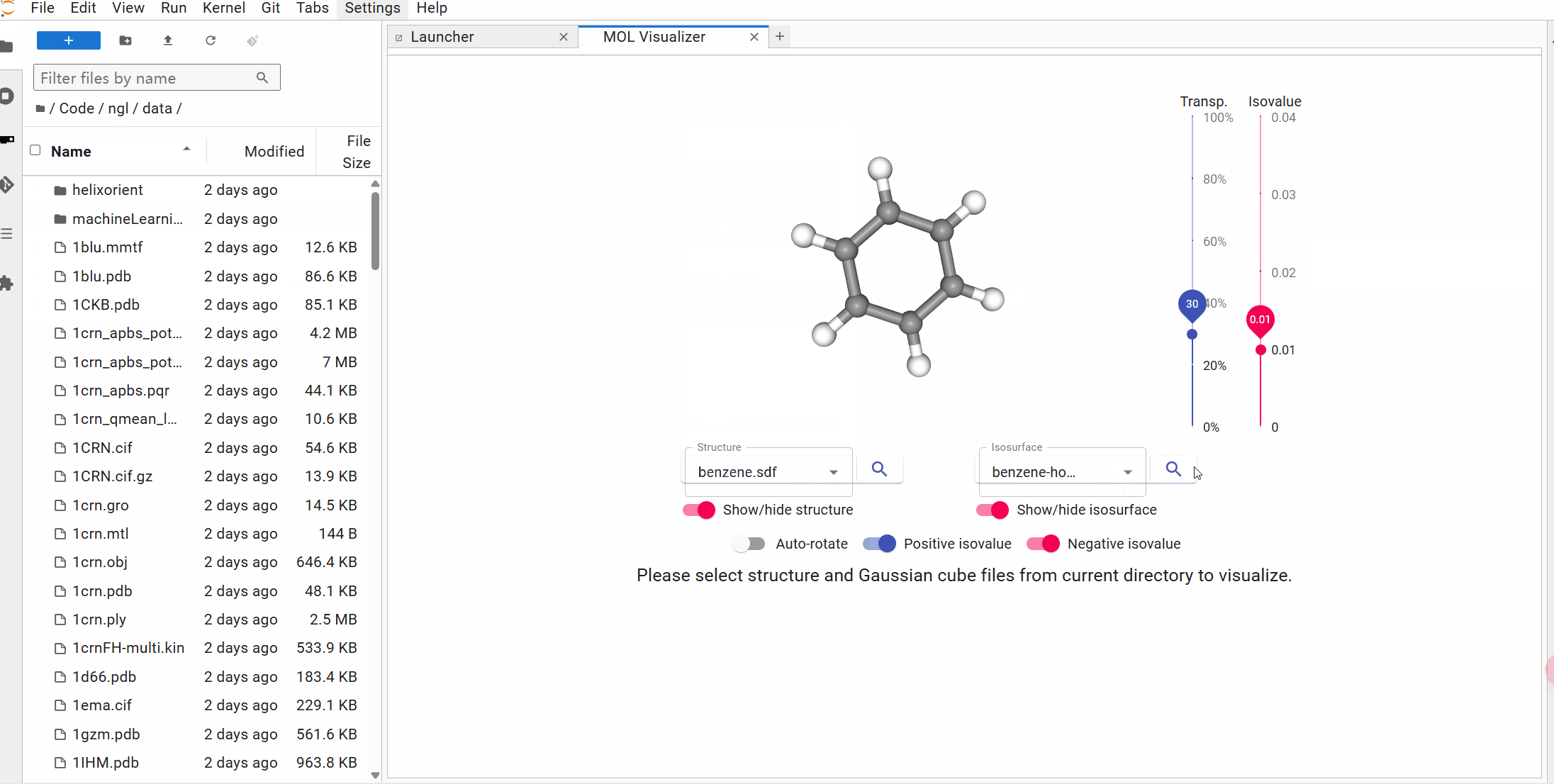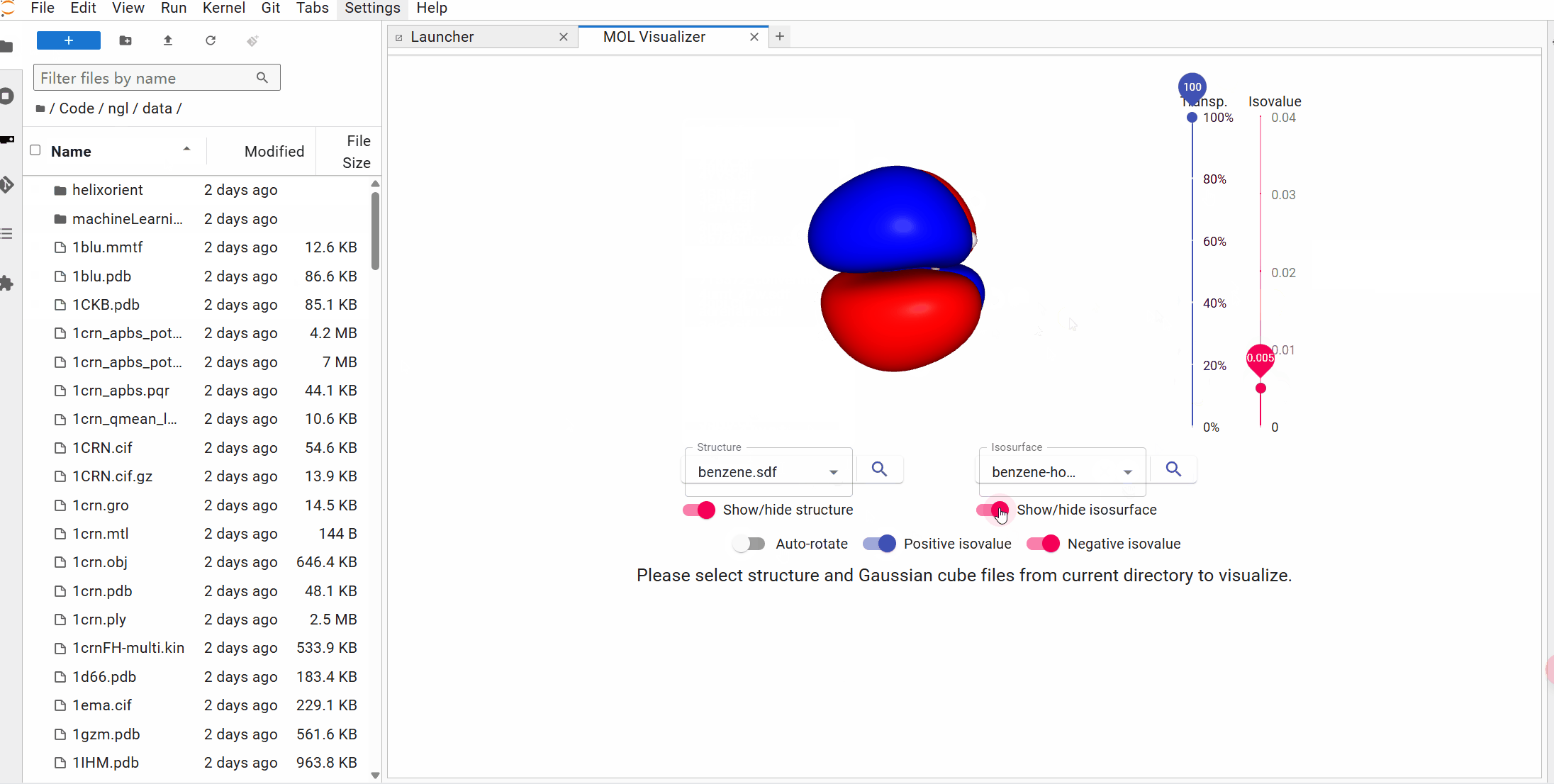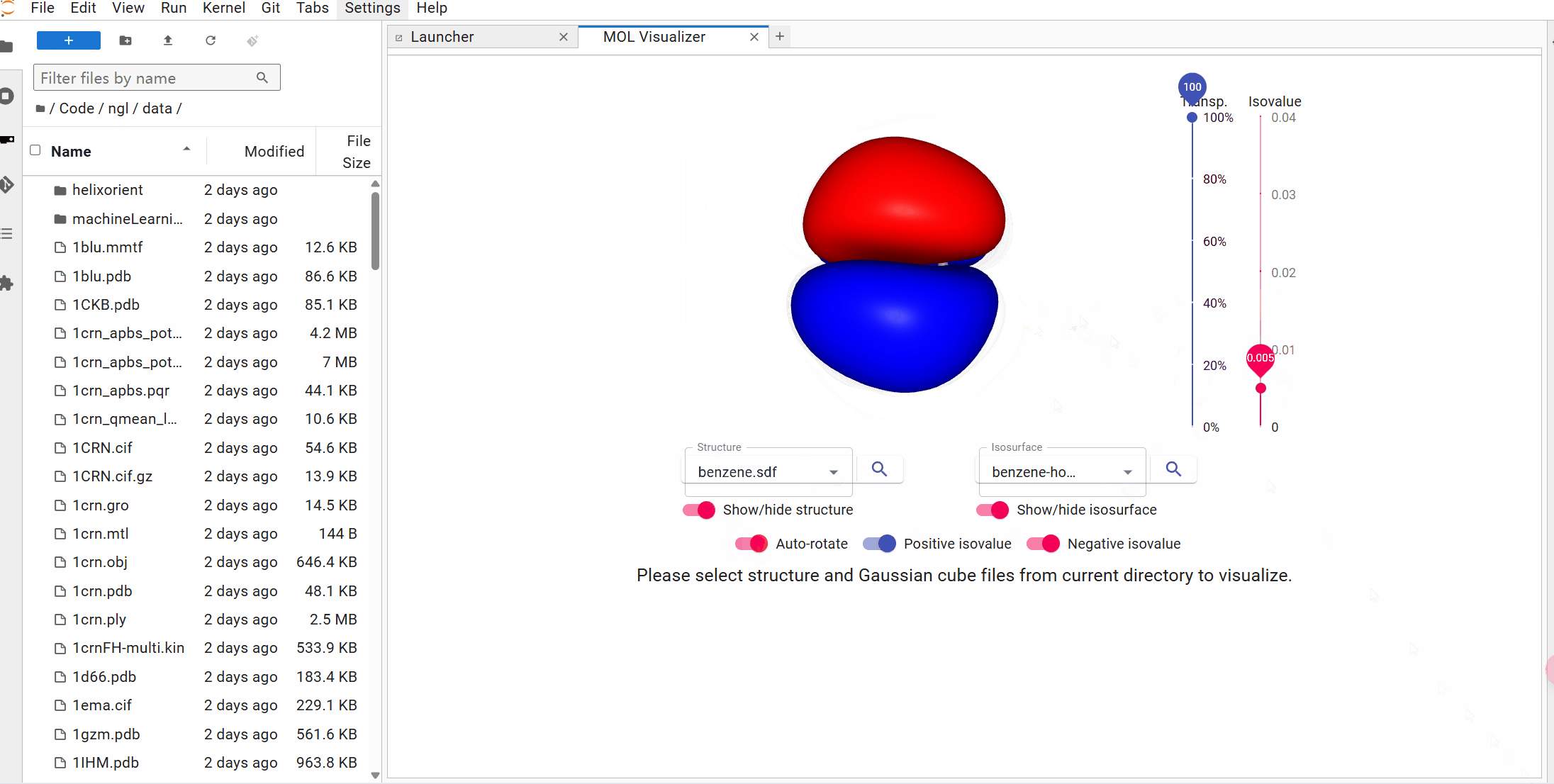
## Try it with Binder
[](https://mybinder.org/v2/gh/osscar-org/jupyterlab-mol-visualizer/main?urlpath=lab)
## Requirements
- JupyterLab >= 4.0.0
## Install
To install the extension, execute:
```bash
pip install jupyterlab_mol_visualizer
```
## Uninstall
To remove the extension, execute:
```bash
pip uninstall jupyterlab_mol_visualizer
```
## Contributing
### Development install
Note: You will need NodeJS to build the extension package.
The `jlpm` command is JupyterLab's pinned version of
[yarn](https://yarnpkg.com/) that is installed with JupyterLab. You may use
`yarn` or `npm` in lieu of `jlpm` below.
```bash
# Clone the repo to your local environment
# Change directory to the jupyterlab_mol_visualizer directory
# Install package in development mode
pip install -e "."
# Link your development version of the extension with JupyterLab
jupyter labextension develop . --overwrite
# Rebuild extension Typescript source after making changes
jlpm build
```
You can watch the source directory and run JupyterLab at the same time in different terminals to watch for changes in the extension's source and automatically rebuild the extension.
```bash
# Watch the source directory in one terminal, automatically rebuilding when needed
jlpm watch
# Run JupyterLab in another terminal
jupyter lab
```
With the watch command running, every saved change will immediately be built locally and available in your running JupyterLab. Refresh JupyterLab to load the change in your browser (you may need to wait several seconds for the extension to be rebuilt).
By default, the `jlpm build` command generates the source maps for this extension to make it easier to debug using the browser dev tools. To also generate source maps for the JupyterLab core extensions, you can run the following command:
```bash
jupyter lab build --minimize=False
```
### Development uninstall
```bash
pip uninstall jupyterlab_mol_visualizer
```
In development mode, you will also need to remove the symlink created by `jupyter labextension develop`
command. To find its location, you can run `jupyter labextension list` to figure out where the `labextensions`
folder is located. Then you can remove the symlink named `jupyterlab_mol_visualizer` within that folder.
### Testing the extension
#### Frontend tests
This extension is using [Jest](https://jestjs.io/) for JavaScript code testing.
To execute them, execute:
```sh
jlpm
jlpm test
```
#### Integration tests
This extension uses [Playwright](https://playwright.dev) for the integration tests (aka user level tests).
More precisely, the JupyterLab helper [Galata](https://github.com/jupyterlab/jupyterlab/tree/master/galata) is used to handle testing the extension in JupyterLab.
More information are provided within the [ui-tests](./ui-tests/README.md) README.
### Packaging the extension
See [RELEASE](RELEASE.md)
## How to cite
When using the content of this repository, please cite the following two articles:
1. D. Du, T. J. Baird, S. Bonella and G. Pizzi, OSSCAR, an open platform for collaborative development of computational tools for education in science, *Computer Physics Communications*, **282**, 108546 (2023).
https://doi.org/10.1016/j.cpc.2022.108546
2. D. Du, T. J. Baird, K. Eimre, S. Bonella, G. Pizzi, Jupyter widgets and extensions for education and research in computational physics and chemistry, *Computer Physics Communications*, **305**, 109353 (2024).
https://doi.org/10.1016/j.cpc.2024.109353
## Acknowledgements
We acknowledge support from the EPFL Open Science Fund via the [OSSCAR](http://www.osscar.org) project.
