Ecosyste.ms: Awesome
An open API service indexing awesome lists of open source software.
https://github.com/ramiromagno/gwasrapidd
gwasrapidd: an R package to query, download and wrangle GWAS Catalog data
https://github.com/ramiromagno/gwasrapidd
association-studies gwas-catalog human r rest-client snp trait trait-ontology
Last synced: 9 days ago
JSON representation
gwasrapidd: an R package to query, download and wrangle GWAS Catalog data
- Host: GitHub
- URL: https://github.com/ramiromagno/gwasrapidd
- Owner: ramiromagno
- License: other
- Created: 2018-10-31T11:55:20.000Z (over 6 years ago)
- Default Branch: master
- Last Pushed: 2023-12-15T16:36:49.000Z (about 1 year ago)
- Last Synced: 2025-01-16T22:08:51.918Z (16 days ago)
- Topics: association-studies, gwas-catalog, human, r, rest-client, snp, trait, trait-ontology
- Language: R
- Homepage: https://rmagno.eu/gwasrapidd/
- Size: 19.9 MB
- Stars: 95
- Watchers: 4
- Forks: 14
- Open Issues: 5
-
Metadata Files:
- Readme: README.Rmd
- Contributing: .github/CONTRIBUTING.md
- License: LICENSE
- Code of conduct: CODE_OF_CONDUCT.md
Awesome Lists containing this project
README
---
output: github_document
---
```{r setup, include = FALSE}
knitr::opts_chunk$set(
collapse = TRUE,
# comment = "#>",
fig.path = "man/figures/README-",
out.width = "100%"
)
```
# gwasrapidd 
[](https://CRAN.R-project.org/package=gwasrapidd)
[](https://doi.org/10.1093/bioinformatics/btz605)
[](https://www.altmetric.com/details/64505748)
[](https://github.com/ramiromagno/gwasrapidd/actions)
[](https://app.codecov.io/gh/ramiromagno/gwasrapidd?branch=master)
[](https://opensource.org/license/mit/)
The goal of `{gwasrapidd}` is to provide programmatic access to the [NHGRI-EBI
Catalog](https://www.ebi.ac.uk/gwas) of published genome-wide association
studies.
Get started by reading the
[documentation](https://rmagno.eu/gwasrapidd/articles/gwasrapidd.html).
## Installation
Install `{gwasrapidd}` from CRAN:
``` r
install.packages("gwasrapidd")
```
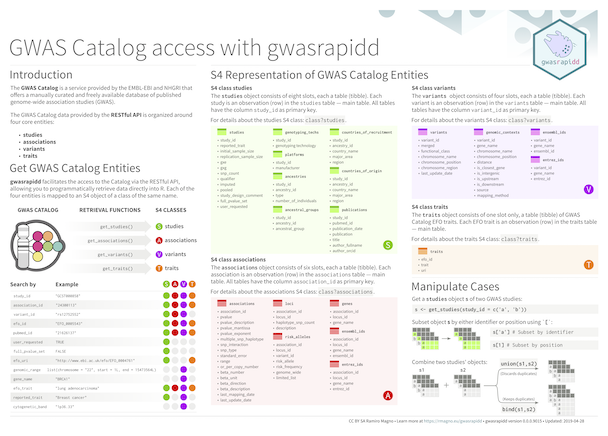
## Cheatsheet
## Example
Get studies related to triple-negative breast cancer:
```{r example1}
library(gwasrapidd)
studies <- get_studies(efo_trait = 'triple-negative breast cancer')
studies@studies[1:4]
```
Find associated variants with study `r studies@studies$study_id[1]`:
```{r example2}
variants <- get_variants(study_id = 'GCST002305')
variants@variants[c('variant_id', 'functional_class')]
```
## Citing this work
`{gwasrapidd}` was published in Bioinformatics in 2019:
https://doi.org/10.1093/bioinformatics/btz605.
To generate a citation for this publication from within R:
```{r citation}
citation('gwasrapidd')
```
## Code of Conduct
Please note that the `{gwasrapidd}` project is released with a [Contributor Code
of Conduct](https://rmagno.eu/gwasrapidd/CODE_OF_CONDUCT.html). By contributing
to this project, you agree to abide by its terms.
## Similar projects
- Bioconductor R package *gwascat* by Vincent J Carey:
[https://www.bioconductor.org/packages/release/bioc/html/gwascat.html](https://www.bioconductor.org/packages/release/bioc/html/gwascat.html)
- Web application *PhenoScanner V2* by Mihir A. Kamat, James R. Staley, and
others:
[http://www.phenoscanner.medschl.cam.ac.uk/](http://www.phenoscanner.medschl.cam.ac.uk/)
- Web application *GWEHS: Genome-Wide Effect sizes and Heritability Screener* by
Eugenio López-Cortegano and Armando Caballero:
[http://gwehs.uvigo.es/](http://gwehs.uvigo.es/)
## Acknowledgements
This work would have not been possible without the precious help from the GWAS
Catalog team, particularly [Daniel
Suveges](https://www.ebi.ac.uk/about/people/daniel-suveges).