https://github.com/rcedgar/muscle
Multiple sequence and structure alignment with top benchmark scores scalable to thousands of sequences. Generates replicate alignments, enabling assessment of downstream analyses such as trees and predicted structures.
https://github.com/rcedgar/muscle
algorithms bioinformatics biology nucleotide-alignment protein-alignment protein-structure protein-structure-alignment sequence-clustering sequence-search
Last synced: 6 months ago
JSON representation
Multiple sequence and structure alignment with top benchmark scores scalable to thousands of sequences. Generates replicate alignments, enabling assessment of downstream analyses such as trees and predicted structures.
- Host: GitHub
- URL: https://github.com/rcedgar/muscle
- Owner: rcedgar
- License: gpl-3.0
- Created: 2021-06-19T21:32:31.000Z (over 4 years ago)
- Default Branch: main
- Last Pushed: 2025-04-12T14:20:09.000Z (6 months ago)
- Last Synced: 2025-04-12T15:27:59.636Z (6 months ago)
- Topics: algorithms, bioinformatics, biology, nucleotide-alignment, protein-alignment, protein-structure, protein-structure-alignment, sequence-clustering, sequence-search
- Language: C++
- Homepage: https://drive5.com/muscle
- Size: 67.8 MB
- Stars: 240
- Watchers: 6
- Forks: 25
- Open Issues: 7
-
Metadata Files:
- Readme: README.md
- Contributing: CONTRIBUTING.md
- License: LICENSE
Awesome Lists containing this project
README
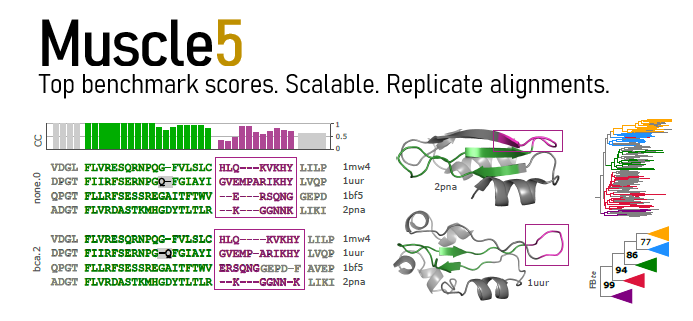
Muscle is widely-used software for making multiple alignments of biological sequences.
Muscle achieves highest scores on Balibase, Bralibase and Balifam benchmark tests and scales to thousands of sequences or structures on a commodity desktop computer.
Muscle supports generating an ensemble of alternative alignments with the same high accuracy obtained with default parameters. By comparing downstream predictions from different alignments, such as trees, a biologist can evaluation the robustness of conclusions against alignment variation caused by ambiguities and errors.
### Multiple structure alignment
Structure alignment ("Muscle-3D") is supported as well as conventional amino acid sequence alignment. Input for structure alignment is a "mega" file generated by the `pdb2mega` command of `reseek` (https://github.com/rcedgar/reseek).
[ ](https://www.youtube.com/watch?v=BzIgqdm9xDs)
](https://www.youtube.com/watch?v=BzIgqdm9xDs)
# for up to ~100 structures
reseek -pdb2mega STRUCTS -output structs.mega
muscle -align structs.mega -output structs.afa
# for up to ~10,000 structures
reseek -convert STRUCTS -bca structs.bca
reseek -pdb2mega structs.bca -output structs.mega
reseek -distmx structs.bca -output structs.distmx
muscle -super7 structs.mega -distmxin structs.distmx -reseek -output structs.afa
### Downloads and installation
Binary files are self-contained, no dependencies. To install, download the binary and make sure the execute bit is set.
https://github.com/rcedgar/muscle/releases
### Documentation
[Muscle v5 home page](https://drive5.com/muscle5)
[Manual](https://drive5.com/muscle5/manual)
### Building MUSCLE from source
[https://github.com/rcedgar/muscle/wiki/Building-MUSCLE](https://github.com/rcedgar/muscle/wiki/Building-MUSCLE)
### References
Edgar RC., Muscle5: High-accuracy alignment ensembles enable unbiased assessments of sequence homology and phylogeny. Nature Communications 13.1 (2022): 6968.
[https://www.nature.com/articles/s41467-022-34630-w.pdf](https://www.nature.com/articles/s41467-022-34630-w.pdf)
Edgar RC. and Tolstoy I., Muscle-3D: scalable multiple protein structure alignment (2024) BioRxiv.