https://github.com/rosettacommons/protein_generator
Joint sequence and structure generation with RoseTTAFold sequence space diffusion
https://github.com/rosettacommons/protein_generator
Last synced: 6 months ago
JSON representation
Joint sequence and structure generation with RoseTTAFold sequence space diffusion
- Host: GitHub
- URL: https://github.com/rosettacommons/protein_generator
- Owner: RosettaCommons
- License: mit
- Created: 2023-05-05T23:24:09.000Z (over 2 years ago)
- Default Branch: main
- Last Pushed: 2024-10-28T21:11:02.000Z (about 1 year ago)
- Last Synced: 2025-04-12T04:49:19.266Z (7 months ago)
- Language: Python
- Homepage: https://huggingface.co/spaces/merle/PROTEIN_GENERATOR
- Size: 50.8 MB
- Stars: 310
- Watchers: 13
- Forks: 52
- Open Issues: 14
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
README
# ProteinGenerator: Generate sequence-structure pairs with RoseTTAFold
This is the Github repository for the [PROTEIN GENERATOR PAPER](https://www.nature.com/articles/s41587-024-02395-w)

## Getting Started
The easiest way to get started is with [PROTEIN GENERATOR](https://huggingface.co/spaces/merle/PROTEIN_GENERATOR) a HuggingFace space where you can play around with the model!
Before running inference you will need to set up a custom conda environment.
Start by creating a new conda environment using the environment.yml file provided in the repository
conda env create -f environment.yml and activating it source activate proteingenerator. Please make sure to modify the CUDA version and dgl version accordingly. Please refer to the [dgl website](https://www.dgl.ai/pages/start.html) for more information.
Once everything has been installed you can download checkpoints:
- [base checkpoint](http://files.ipd.uw.edu/pub/sequence_diffusion/checkpoints/SEQDIFF_221219_equalTASKS_nostrSELFCOND_mod30.pt)
- [DSSP + hotspot checkpoint](http://files.ipd.uw.edu/pub/sequence_diffusion/checkpoints/SEQDIFF_230205_dssp_hotspots_25mask_EQtasks_mod30.pt)
The easiest way to get started is opening the protein_generator.ipynb notebook and running the sampler class interactively, when ready to submit a production run use the output agrs.json file to launch:
python ./inference.py -input_json ./examples/out/design_000000_args.json
\* note that to get the notebook running you will need to add the custom conda environment as a jupyter kernel, see how to do this [here](https://towardsdatascience.com/get-your-conda-environment-to-show-in-jupyter-notebooks-the-easy-way-17010b76e874)
Check out the templates in the [example folder](examples) to see how you can set up jobs for the various design strategies
## Adding new sequence based potentials
To add a custom potential to guide the sequence diffusion process toward your desired space, you can add potentials into utils/potentials.py. At the top of the file a template class is provided with functions that are required to implement your potential. It can be helpful to look through the other potentials in this file to see examples of how to implement. At the bottom of the file is a dictionary mapping the name used in the potentials argument to the class name in file.
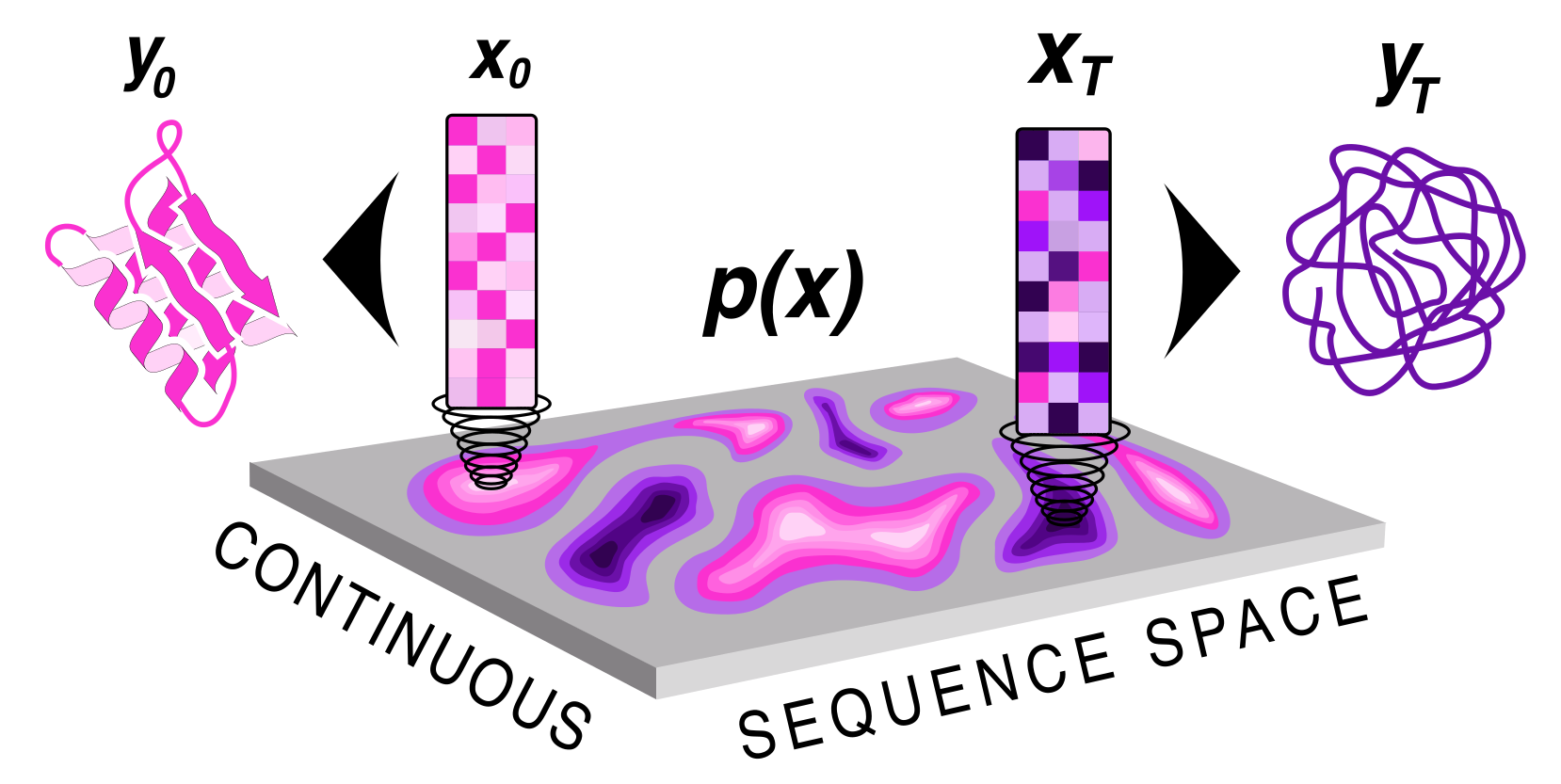
## About the model
ProteinGenerator is trained on the same dataset and uses the same architecture as RoseTTAFold. To train the model, a ground truth sequence is transformed into an Lx20 continuous space and gaussian noise is added to diffuse the sequence to the sampled timestep. To condition on structure and sequence, the structre for a motif is given and then corresponding sequence is denoised in the input. The rest of the structure is blackhole initialized. For each example the model is trained to predict Xo and losses are applied on the structure and sequence respectively. During training big T is set to 1000 steps, and a square root schedule is used to add noise.
## Looking ahead
We are excited for the community to get involved writing new potentials and building out the codebase further!
## Acknowledgements
We would like to thank Frank DiMaio and Minkyung Baek who developed RoseTTAFold which allowed us to build out this platform. Other acknowledgements for code and development please see the preprint.