https://github.com/rusty1s/himp-gnn
Hierarchical Inter-Message Passing for Learning on Molecular Graphs
https://github.com/rusty1s/himp-gnn
geometric-deep-learning graph-neural-networks graph-pooling junction-tree molecular-graph pytorch
Last synced: about 1 month ago
JSON representation
Hierarchical Inter-Message Passing for Learning on Molecular Graphs
- Host: GitHub
- URL: https://github.com/rusty1s/himp-gnn
- Owner: rusty1s
- Created: 2020-06-22T11:48:01.000Z (almost 5 years ago)
- Default Branch: master
- Last Pushed: 2021-12-07T09:48:13.000Z (over 3 years ago)
- Last Synced: 2024-11-29T18:39:54.326Z (6 months ago)
- Topics: geometric-deep-learning, graph-neural-networks, graph-pooling, junction-tree, molecular-graph, pytorch
- Language: Python
- Homepage: https://arxiv.org/abs/2006.12179
- Size: 163 KB
- Stars: 77
- Watchers: 5
- Forks: 6
- Open Issues: 5
-
Metadata Files:
- Readme: README.md
Awesome Lists containing this project
README
Hierarchical Inter-Message Passing for Learning on Molecular Graphs
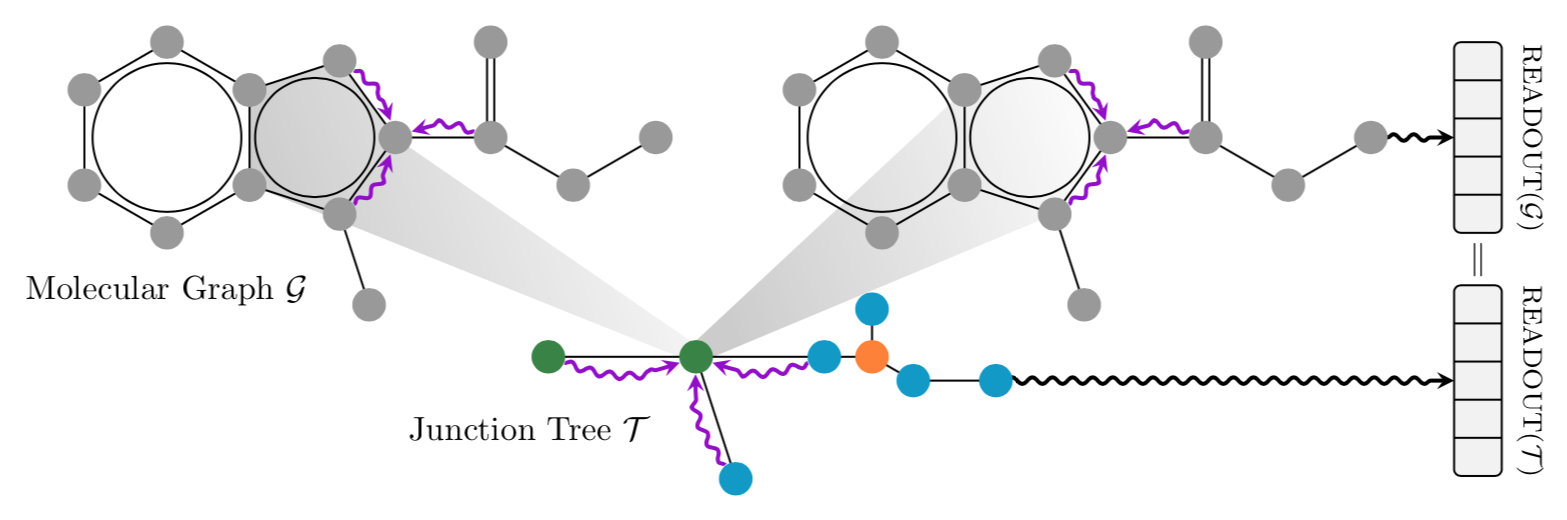
--------------------------------------------------------------------------------
This is a PyTorch implementation of **Hierarchical Inter-Message Passing for Learning on Molecular Graphs**, as described in our paper:
Matthias Fey, Jan-Gin Yuen, Frank Weichert: [Hierarchical Inter-Message Passing for Learning on Molecular Graphs](https://arxiv.org/abs/2006.12179) *(GRL+ 2020)*
## Requirements
* **[PyTorch](https://pytorch.org/get-started/locally/)** (>=1.4.0)
* **[PyTorch Geometric](https://github.com/rusty1s/pytorch_geometric)** (>=1.5.0)
* **[OGB](https://ogb.stanford.edu/)** (>=1.1.0)
## Experiments
Experiments can be run via:
```
$ python train_zinc_subset.py
$ python train_zinc_full.py
$ python train_hiv.py
$ python train_muv.py
$ python train_tox21.py
$ python train_ogbhiv.py
$ python train_ogbpcba.py
```
## Cite
Please cite [our paper](https://arxiv.org/abs/2006.12179) if you use this code in your own work:
```
@inproceedings{Fey/etal/2020,
title={Hierarchical Inter-Message Passing for Learning on Molecular Graphs},
author={Fey, M. and Yuen, J. G. and Weichert, F.},
booktitle={ICML Graph Representation Learning and Beyond (GRL+) Workhop},
year={2020},
}
```