https://github.com/saezlab/decoupler
R package to infer biological activities from omics data using a collection of methods.
https://github.com/saezlab/decoupler
r r-package rstats
Last synced: 3 months ago
JSON representation
R package to infer biological activities from omics data using a collection of methods.
- Host: GitHub
- URL: https://github.com/saezlab/decoupler
- Owner: saezlab
- License: gpl-3.0
- Created: 2019-11-21T21:51:19.000Z (over 5 years ago)
- Default Branch: master
- Last Pushed: 2025-03-12T08:23:48.000Z (4 months ago)
- Last Synced: 2025-04-03T04:08:09.954Z (3 months ago)
- Topics: r, r-package, rstats
- Language: R
- Homepage: https://saezlab.github.io/decoupleR/
- Size: 38.5 MB
- Stars: 235
- Watchers: 4
- Forks: 25
- Open Issues: 3
-
Metadata Files:
- Readme: README.Rmd
- Changelog: NEWS.md
- License: LICENSE
Awesome Lists containing this project
README
---
output: github_document
---
```{r, include = FALSE}
knitr::opts_chunk$set(
collapse = TRUE,
comment = "#>",
fig.path = "man/figures/README-",
out.width = "100%"
)
```
# decoupleR 
[](https://www.tidyverse.org/lifecycle/#maturing)
[](https://bioconductor.org/checkResults/release/bioc-LATEST/decoupleR)
[](https://bioconductor.org/checkResults/devel/bioc-LATEST/decoupleR)
[](https://github.com/saezlab/decoupleR/actions)
[](https://codecov.io/gh/saezlab/decoupleR?branch=master)
[](https://github.com/saezlab/decoupleR/issues)
## Overview
There are many methods that allow us to extract biological activities from omics data.
`decoupleR` is a Bioconductor package containing different statistical methods to
extract biological signatures from prior knowledge within a unified framework.
Additionally, it incorporates methods that take into account the sign and weight of
network interactions. `decoupleR` can be used with any omic, as long as its
features can be linked to a biological process based on prior knowledge.
For example, in transcriptomics gene sets regulated by a transcription
factor, or in phospho-proteomics phosphosites that are targeted by a kinase.
This is the R version, for its faster and memory efficient Python implementation go [here](https://decoupler-py.readthedocs.io/en/latest/).
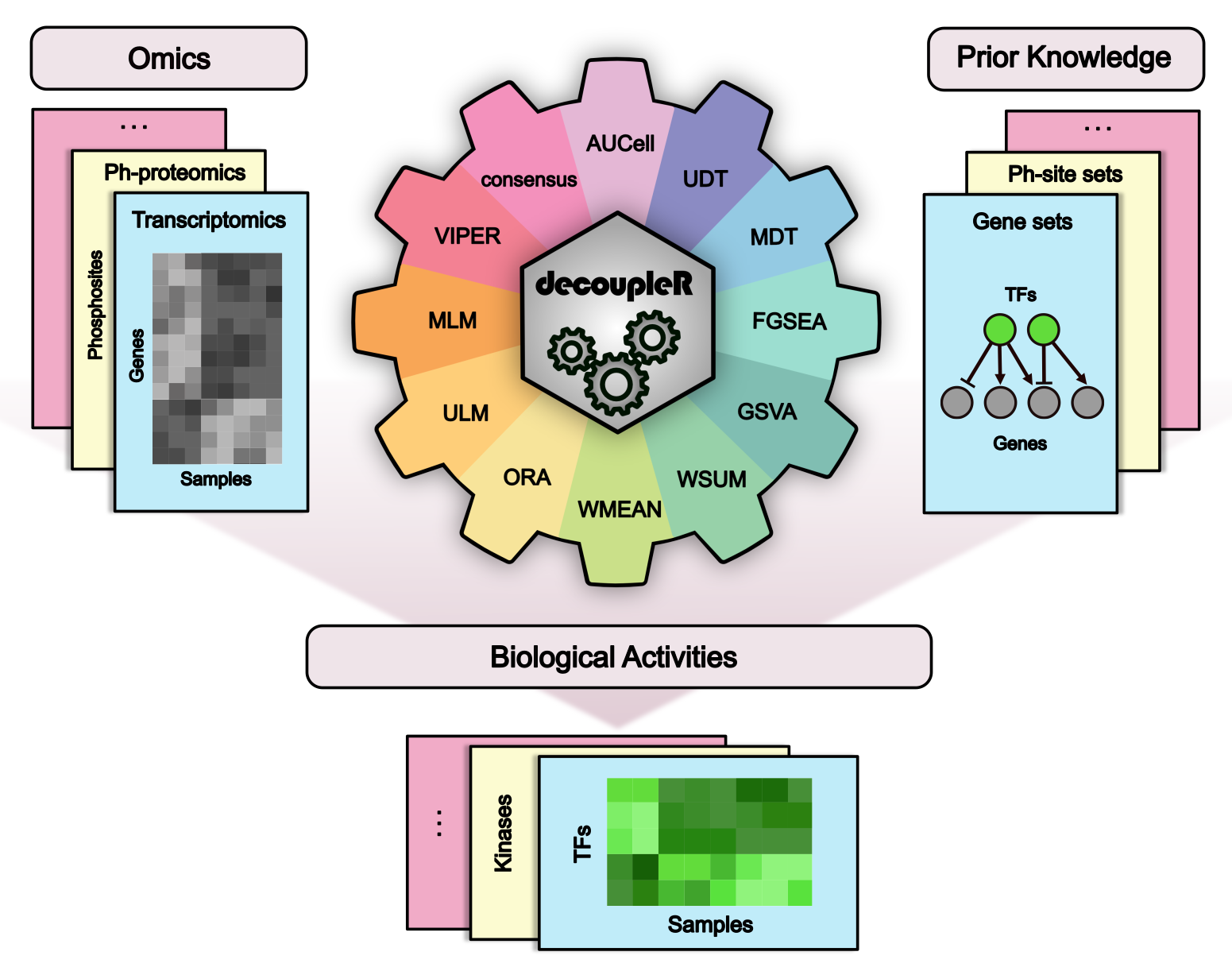
For more information about how this package has been used with real data,
please check the following links:
- [decoupleR's general usage](https://saezlab.github.io/decoupleR/articles/decoupleR.html)
- [Pathway activity inference in bulk RNA-seq](https://saezlab.github.io/decoupleR/articles/pw_bk.html)
- [Pathway activity inference from scRNA-seq](https://saezlab.github.io/decoupleR/articles/pw_sc.html)
- [Transcription factor activity inference in bulk RNA-seq](https://saezlab.github.io/decoupleR/articles/tf_bk.html)
- [Transcription factor activity inference from scRNA-seq](https://saezlab.github.io/decoupleR/articles/tf_sc.html)
- [Example of Kinase and TF activity estimation](https://saezlab.github.io/kinase_tf_mini_tuto/)
- [decoupleR's manuscript repository](https://github.com/saezlab/decoupleR_manuscript)
- [Python implementation](https://decoupler-py.readthedocs.io/en/latest/)
# Installation
`decoupleR` is an R package distributed as part of the Bioconductor
project. To install the package, start R and enter:
```{r bioconductor_install, eval=FALSE}
install.packages("BiocManager")
BiocManager::install("decoupleR")
```
Alternatively, you can instead install the latest development version from [GitHub](https://github.com/) with:
```{r github_install, eval=FALSE}
BiocManager::install("saezlab/decoupleR")
```
## License
Footprint methods inside `decoupleR` can be used for academic or commercial purposes, except `viper` which holds a non-commercial license.
The data redistributed by `OmniPath` does not have a license, each original resource carries their own.
[Here](https://omnipathdb.org/info) one can find the license information of all the resources in `OmniPath`.
## Citation
Badia-i-Mompel P., Vélez Santiago J., Braunger J., Geiss C., Dimitrov D., Müller-Dott S., Taus P., Dugourd A., Holland
C.H., Ramirez Flores R.O. and Saez-Rodriguez J. 2022. decoupleR: ensemble of computational methods to infer
biological activities from omics data. Bioinformatics Advances. https://doi.org/10.1093/bioadv/vbac016