https://github.com/saezlab/liana-py
LIANA+: an all-in-one framework for cell-cell communication
https://github.com/saezlab/liana-py
cell-cell-communication ligand-receptor python single-cell single-cell-rna-seq spatial spatialomics
Last synced: 10 months ago
JSON representation
LIANA+: an all-in-one framework for cell-cell communication
- Host: GitHub
- URL: https://github.com/saezlab/liana-py
- Owner: saezlab
- License: gpl-3.0
- Created: 2022-09-22T13:20:35.000Z (over 3 years ago)
- Default Branch: main
- Last Pushed: 2024-07-18T11:49:07.000Z (over 1 year ago)
- Last Synced: 2024-08-07T13:28:08.758Z (over 1 year ago)
- Topics: cell-cell-communication, ligand-receptor, python, single-cell, single-cell-rna-seq, spatial, spatialomics
- Language: Python
- Homepage: http://liana-py.readthedocs.io/
- Size: 224 MB
- Stars: 141
- Watchers: 2
- Forks: 16
- Open Issues: 27
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
README
# LIANA+: an all-in-one cell-cell communication framework 
[](https://github.com/saezlab/liana-py/actions)
[](https://github.com/saezlab/liana-py/issues/)
[](https://liana-py.readthedocs.io/en/latest/?badge=latest)
[](https://codecov.io/gh/saezlab/liana-py)
[](https://pepy.tech/project/liana)
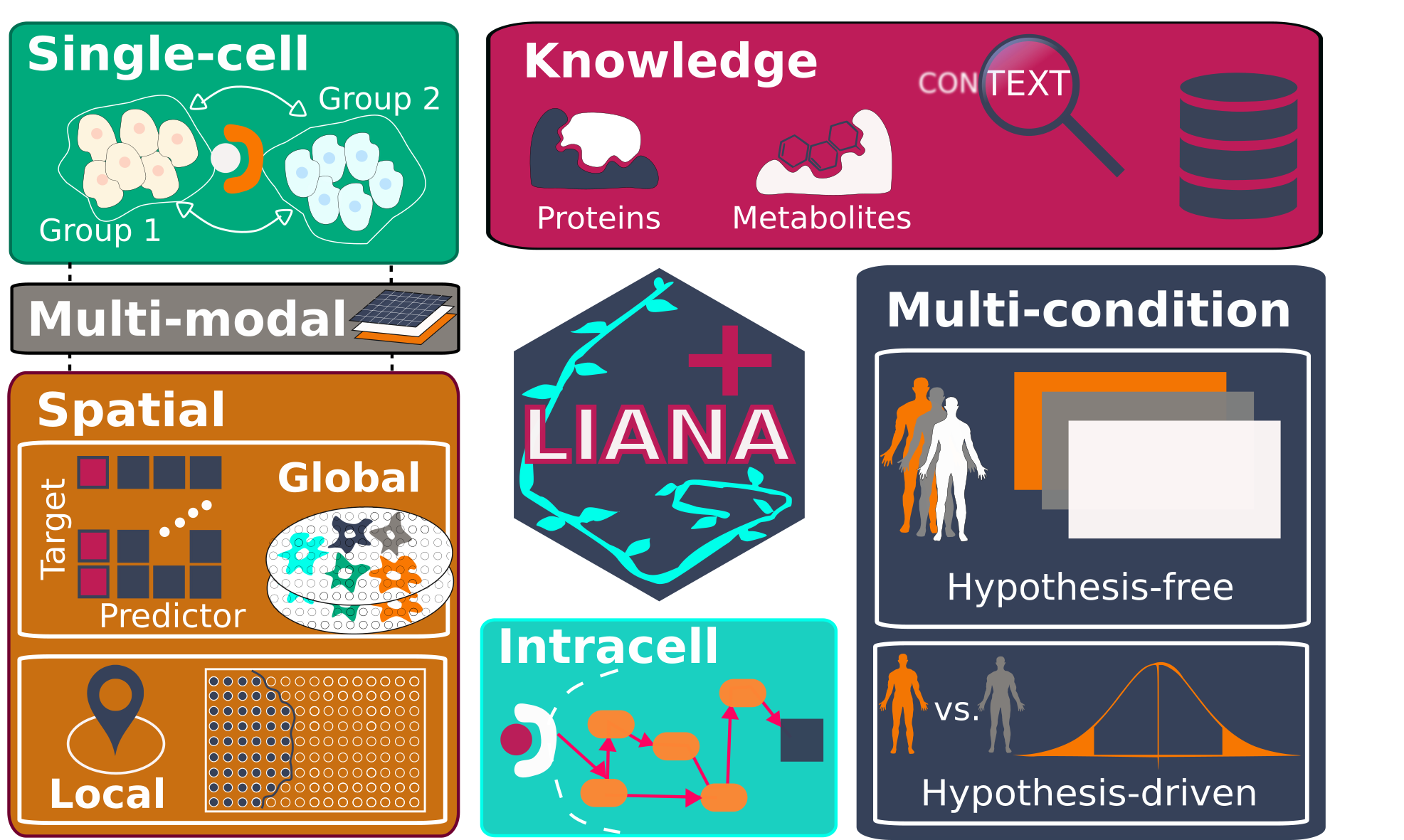
LIANA+ is a scalable framework that adapts and extends existing methods and knowledge to study cell-cell communication in single-cell, spatially-resolved, and multi-modal omics data. It is part of the [scverse ecosystem](https://github.com/scverse), and relies on [AnnData](https://github.com/scverse/anndata) & [MuData](https://github.com/scverse/mudata) objects as input.
## Contributions
We welcome suggestions, ideas, and contributions! Please do not hesitate to contact us, open issues, and check the [contributions guide](https://liana-py.readthedocs.io/en/latest/contributing.html).
## Vignettes
A set of extensive vignettes can be found in the [LIANA+ documentation](https://liana-py.readthedocs.io/en/latest/).
## Decision Tree
### Does the data contain spatial coordinates?
#### Yes
- **Q: Bivariate or unsupervised, multi-variate, and multi-view analysis?**
- **Bivariate:**
- **Q: Are you interested in identifying the subregions of interactions (i.e., local interactions)?**
- **Yes:** Check the [**Local** Bivariate Metrics](https://liana-py.readthedocs.io/en/latest/notebooks/bivariate.html#Bivariate-Ligand-Receptor-Relationships)
- **No:** Check the [**Global** Bivariate Metrics](https://liana-py.readthedocs.io/en/latest/notebooks/bivariate.html#Bivariate-Ligand-Receptor-Relationships)
- **Unsupervised:** [Multi-view learning](https://liana-py.readthedocs.io/en/latest/notebooks/misty.html)
#### No
- **Q: Are you interested in comparing CCC across samples?**
- **Yes:**
- **Q: Are you interested in a specific contrast?**
- **Yes:** [Differential Contrasts and Downstream Signalling](https://liana-py.readthedocs.io/en/latest/notebooks/targeted.html)
- **No:** Unsupervised Cross-conditional LR inference with [MOFA+](https://liana-py.readthedocs.io/en/latest/notebooks/mofatalk.html) or [Tensor-cell2cell](https://liana-py.readthedocs.io/en/latest/notebooks/liana_c2c.html)
- **No:** [Steady-state Ligand-Receptor inference](https://liana-py.readthedocs.io/en/latest/notebooks/basic_usage.html)
### Is your data Multi-modal?
- **Spatial:** [Integrating Multi-Modal Spatially-Resolved Technologies](https://liana-py.readthedocs.io/en/latest/notebooks/sma.html)
- **Non-Spatial:** [Integrating Multi-Modal Single-Cell Technologies](https://liana-py.readthedocs.io/en/latest/notebooks/sc_multi.html)
#### Infer Metabolite-mediated CCC from transcriptomics?
- [Non-spatial Data](https://liana-py.readthedocs.io/en/latest/notebooks/sc_multi.html#Metabolite-mediated-CCC-from-Transcriptomics-Data)
## API
For further information please check LIANA's [API documentation](https://liana-py.readthedocs.io/en/latest/api.html).
## Cite LIANA+:
Dimitrov D., Schäfer P.S.L, Farr E., Rodriguez Mier P., Lobentanzer S., Badia-i-Mompel P., Dugourd A., Tanevski J., Ramirez Flores R.O. and Saez-Rodriguez J. LIANA+ provides an all-in-one framework for cell–cell communication inference. Nat Cell Biol (2024). https://doi.org/10.1038/s41556-024-01469-w
Dimitrov, D., Türei, D., Garrido-Rodriguez M., Burmedi P.L., Nagai, J.S., Boys, C., Flores, R.O.R., Kim, H., Szalai, B., Costa, I.G., Valdeolivas, A., Dugourd, A. and Saez-Rodriguez, J. Comparison of methods and resources for cell-cell communication inference from single-cell RNA-Seq data. Nat Commun 13, 3224 (2022). https://doi.org/10.1038/s41467-022-30755-0
Please also consider citing any of the methods and/or resources that were particularly relevant for your research!