https://github.com/saforem2/llm-workshop-talk
Simple tutorial on creating Small(-ish) LLMs (pt. 2 🎉!!)
https://github.com/saforem2/llm-workshop-talk
Last synced: 8 months ago
JSON representation
Simple tutorial on creating Small(-ish) LLMs (pt. 2 🎉!!)
- Host: GitHub
- URL: https://github.com/saforem2/llm-workshop-talk
- Owner: saforem2
- Created: 2024-02-12T20:58:36.000Z (almost 2 years ago)
- Default Branch: main
- Last Pushed: 2024-09-10T14:51:56.000Z (over 1 year ago)
- Last Synced: 2025-04-23T17:59:05.396Z (9 months ago)
- Language: Python
- Homepage: https://saforem2.github.io/llm-workshop-talk/
- Size: 5.04 MB
- Stars: 3
- Watchers: 1
- Forks: 0
- Open Issues: 0
-
Metadata Files:
- Readme: docs/README 2.html
Awesome Lists containing this project
README
Creating Small(-ish) LLMs
code{white-space: pre-wrap;}
span.smallcaps{font-variant: small-caps;}
div.columns{display: flex; gap: min(4vw, 1.5em);}
div.column{flex: auto; overflow-x: auto;}
div.hanging-indent{margin-left: 1.5em; text-indent: -1.5em;}
ul.task-list{list-style: none;}
ul.task-list li input[type="checkbox"] {
width: 0.8em;
margin: 0 0.8em 0.2em -1em; /* quarto-specific, see https://github.com/quarto-dev/quarto-cli/issues/4556 */
vertical-align: middle;
}
/* CSS for syntax highlighting */
pre > code.sourceCode { white-space: pre; position: relative; }
pre > code.sourceCode > span { line-height: 1.25; }
pre > code.sourceCode > span:empty { height: 1.2em; }
.sourceCode { overflow: visible; }
code.sourceCode > span { color: inherit; text-decoration: inherit; }
div.sourceCode { margin: 1em 0; }
pre.sourceCode { margin: 0; }
@media screen {
div.sourceCode { overflow: auto; }
}
@media print {
pre > code.sourceCode { white-space: pre-wrap; }
pre > code.sourceCode > span { text-indent: -5em; padding-left: 5em; }
}
pre.numberSource code
{ counter-reset: source-line 0; }
pre.numberSource code > span
{ position: relative; left: -4em; counter-increment: source-line; }
pre.numberSource code > span > a:first-child::before
{ content: counter(source-line);
position: relative; left: -1em; text-align: right; vertical-align: baseline;
border: none; display: inline-block;
-webkit-touch-callout: none; -webkit-user-select: none;
-khtml-user-select: none; -moz-user-select: none;
-ms-user-select: none; user-select: none;
padding: 0 4px; width: 4em;
}
pre.numberSource { margin-left: 3em; padding-left: 4px; }
div.sourceCode
{ }
@media screen {
pre > code.sourceCode > span > a:first-child::before { text-decoration: underline; }
}
/* CSS for citations */
div.csl-bib-body { }
div.csl-entry {
clear: both;
margin-bottom: 0em;
}
.hanging-indent div.csl-entry {
margin-left:2em;
text-indent:-2em;
}
div.csl-left-margin {
min-width:2em;
float:left;
}
div.csl-right-inline {
margin-left:2em;
padding-left:1em;
}
div.csl-indent {
margin-left: 2em;
}
{
"location": "navbar",
"copy-button": false,
"collapse-after": 3,
"panel-placement": "end",
"type": "overlay",
"limit": 50,
"keyboard-shortcut": [
"f",
"/",
"s"
],
"show-item-context": false,
"language": {
"search-no-results-text": "No results",
"search-matching-documents-text": "matching documents",
"search-copy-link-title": "Copy link to search",
"search-hide-matches-text": "Hide additional matches",
"search-more-match-text": "more match in this document",
"search-more-matches-text": "more matches in this document",
"search-clear-button-title": "Clear",
"search-text-placeholder": "",
"search-detached-cancel-button-title": "Cancel",
"search-submit-button-title": "Submit",
"search-label": "Search"
}
}
window.dataLayer = window.dataLayer || [];
function gtag(){dataLayer.push(arguments);}
gtag('js', new Date());
gtag('config', 'G-XVM2Y822Y1', { 'anonymize_ip': true});
On this page
- Creating Small(-ish) LLMs
- LLMs from Scratch
- Emergent Abilities
- Training LLMs
- Life-Cycle of the LLM
- Forward Pass
- Generating Text
- Life-Cycle of the LLM: Pre-training
- Life-Cycle of the LLM: Fine-Tuning
- Assistant Models
saforem2/wordplay 🎮💬
saforem2/wordplay 🎮💬
- Install
- Dependencies
- Quick Start
- Model
model.py
- Trainer
trainer.py
- Hands-on Tutorial
- Links
- References
Creating Small(-ish) LLMs
February 13, 2024
February 13, 2024
Creating Small(-ish) LLMs
LLMs from Scratch
Emergent Abilities
Training LLMs
Life-Cycle of the LLM
Forward Pass
Generating Text
Life-Cycle of the LLM: Pre-training
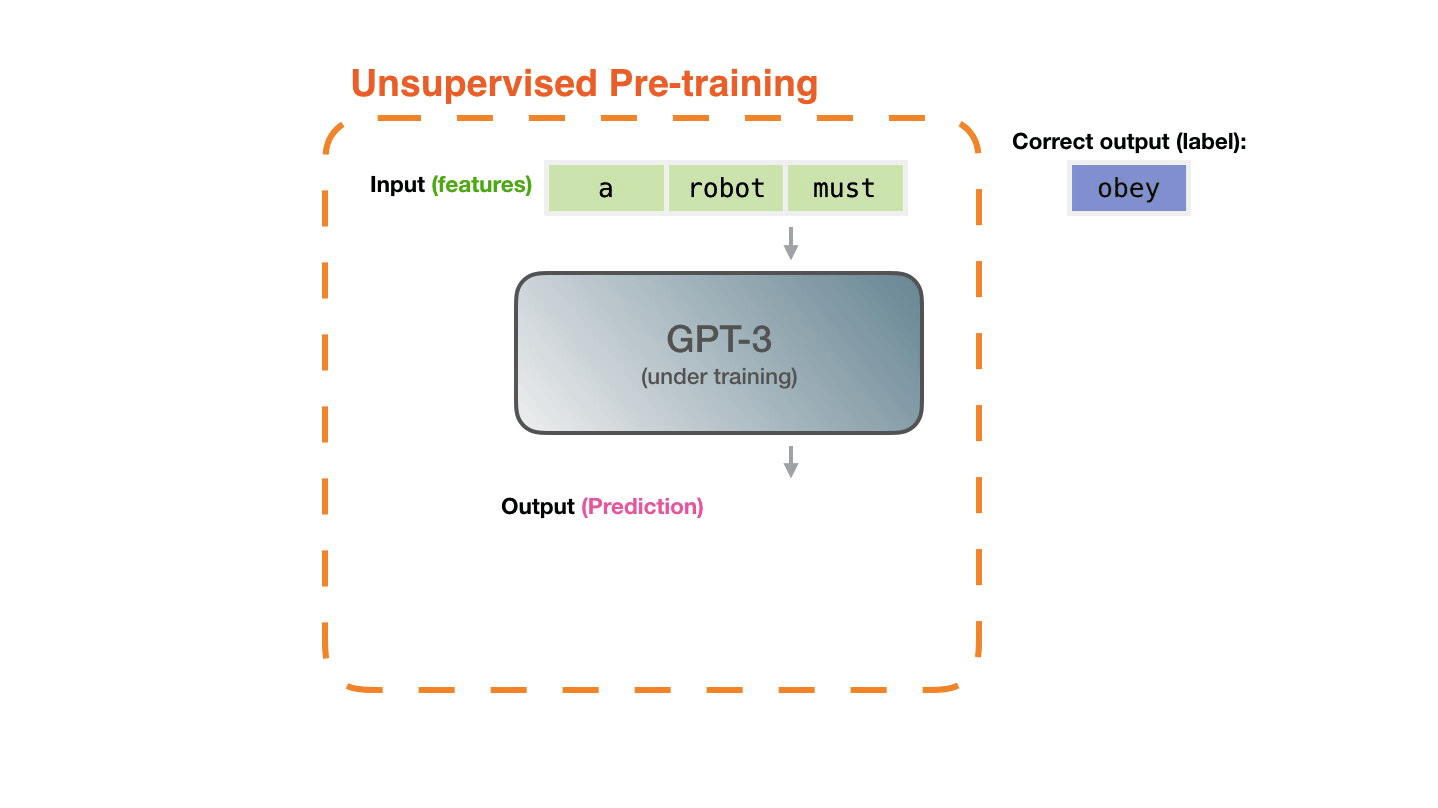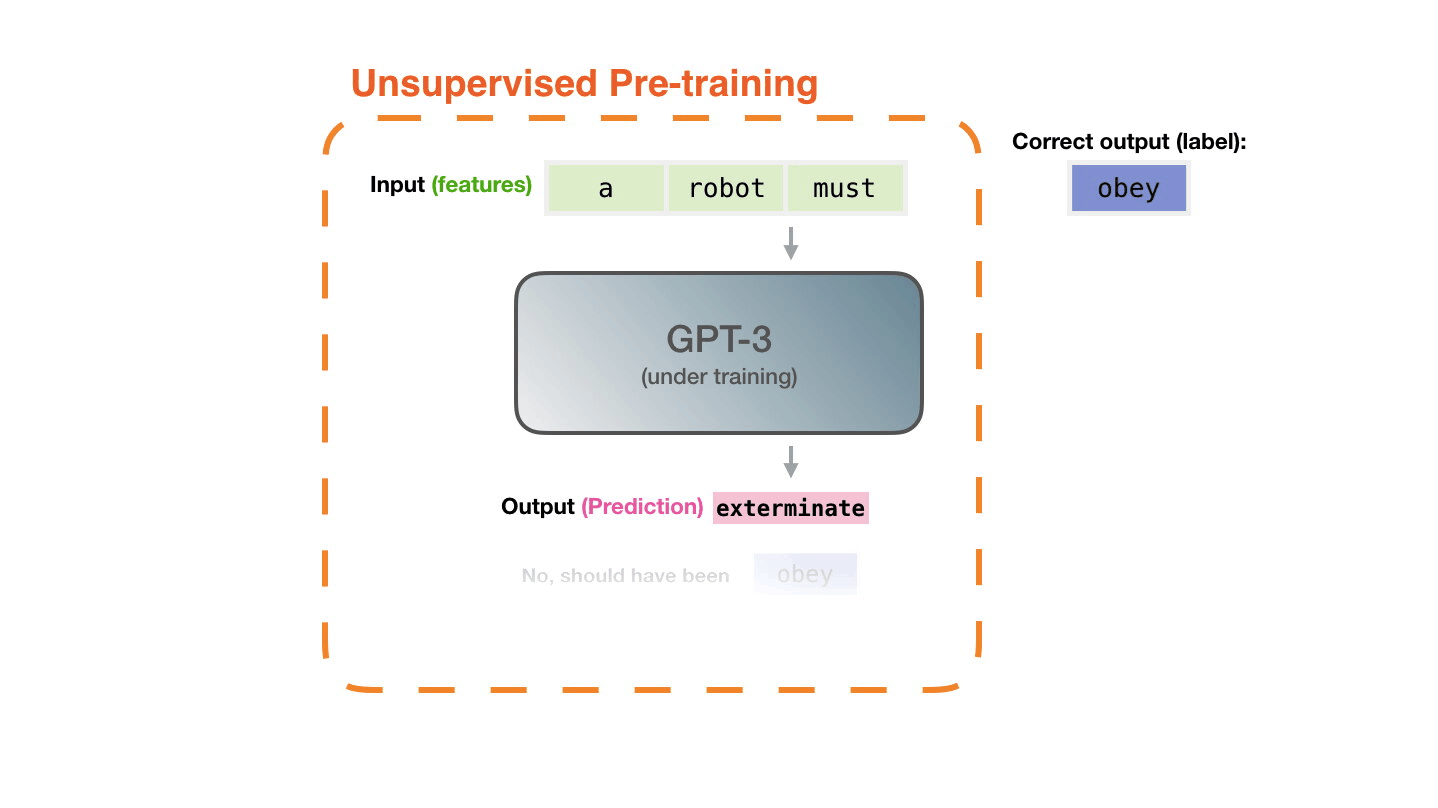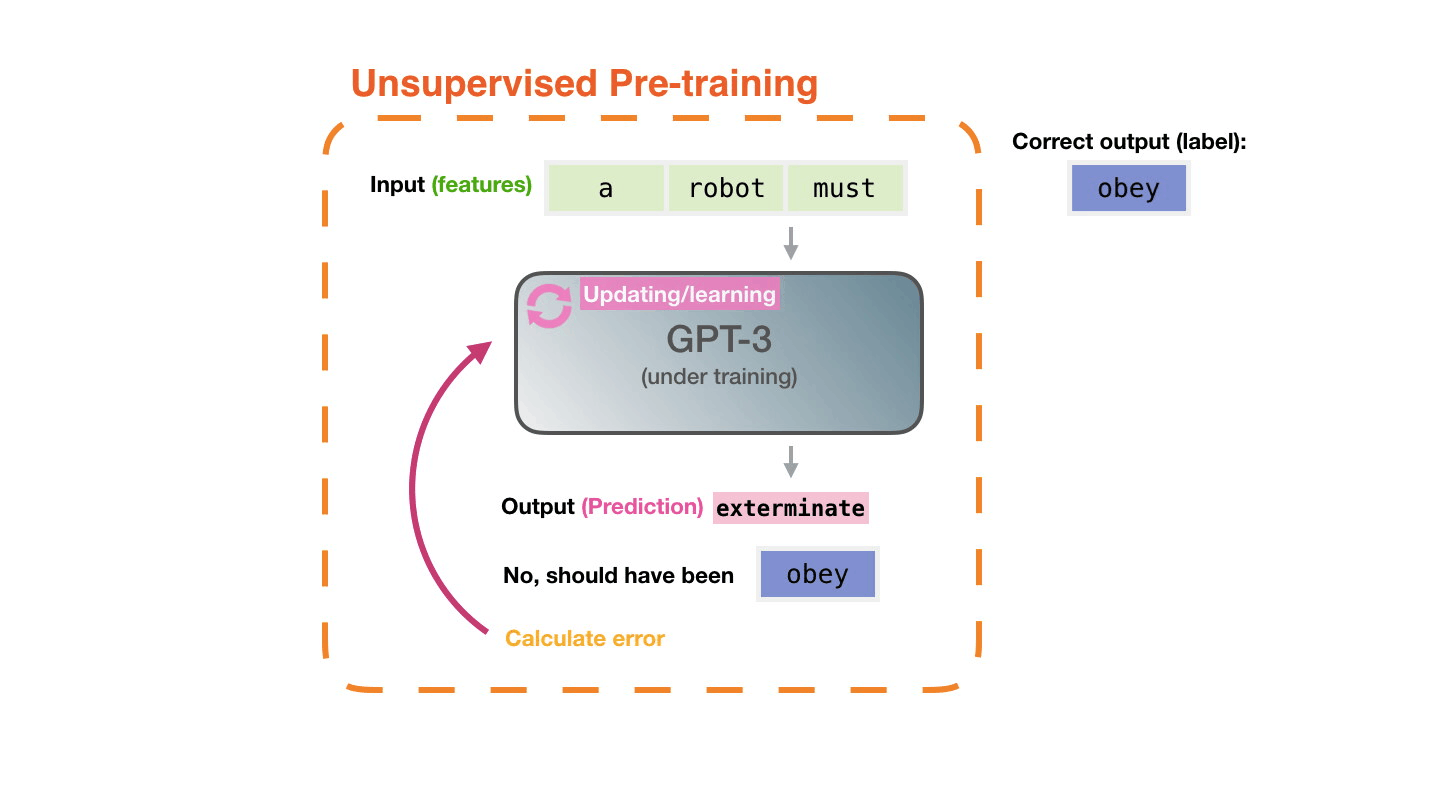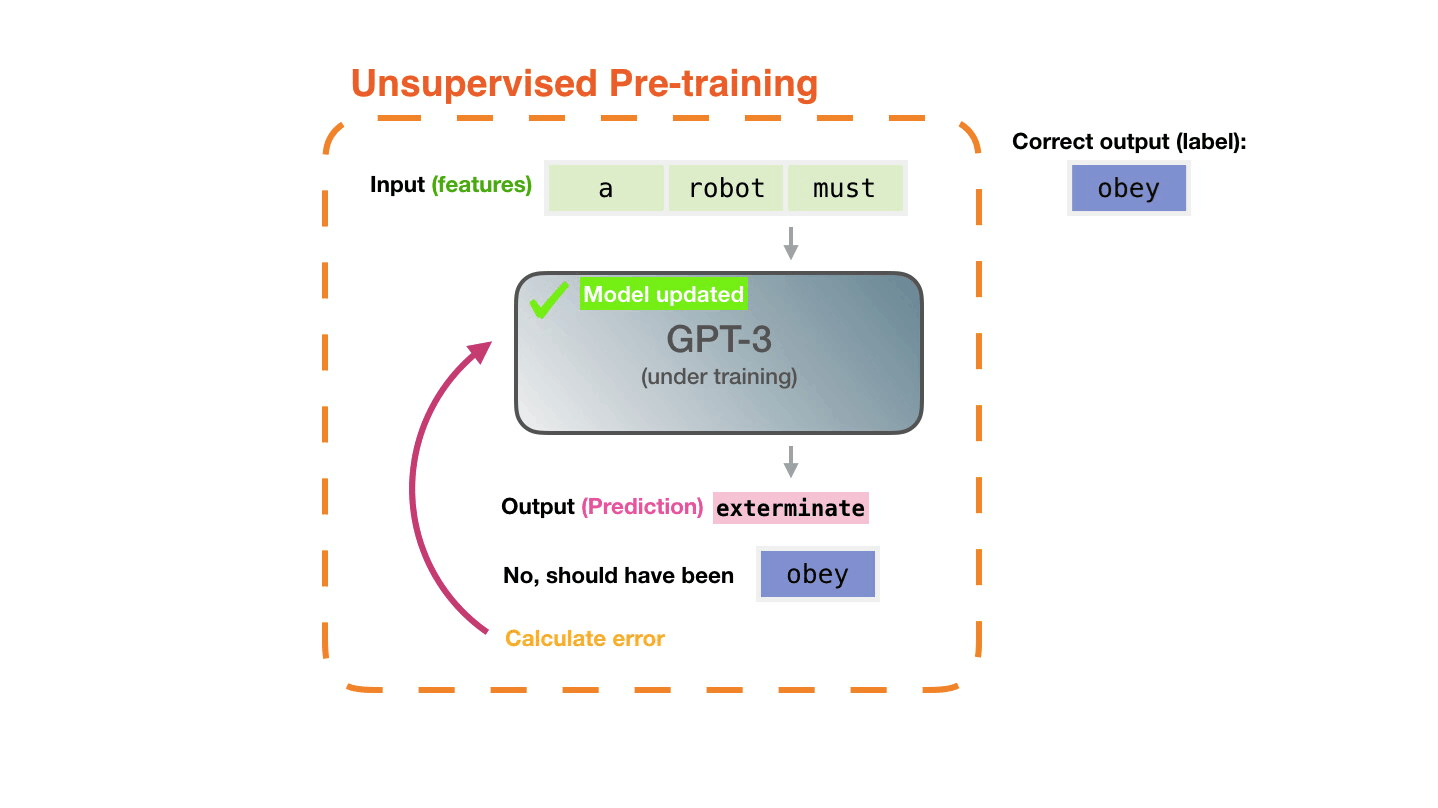
Life-Cycle of the LLM: Fine-Tuning
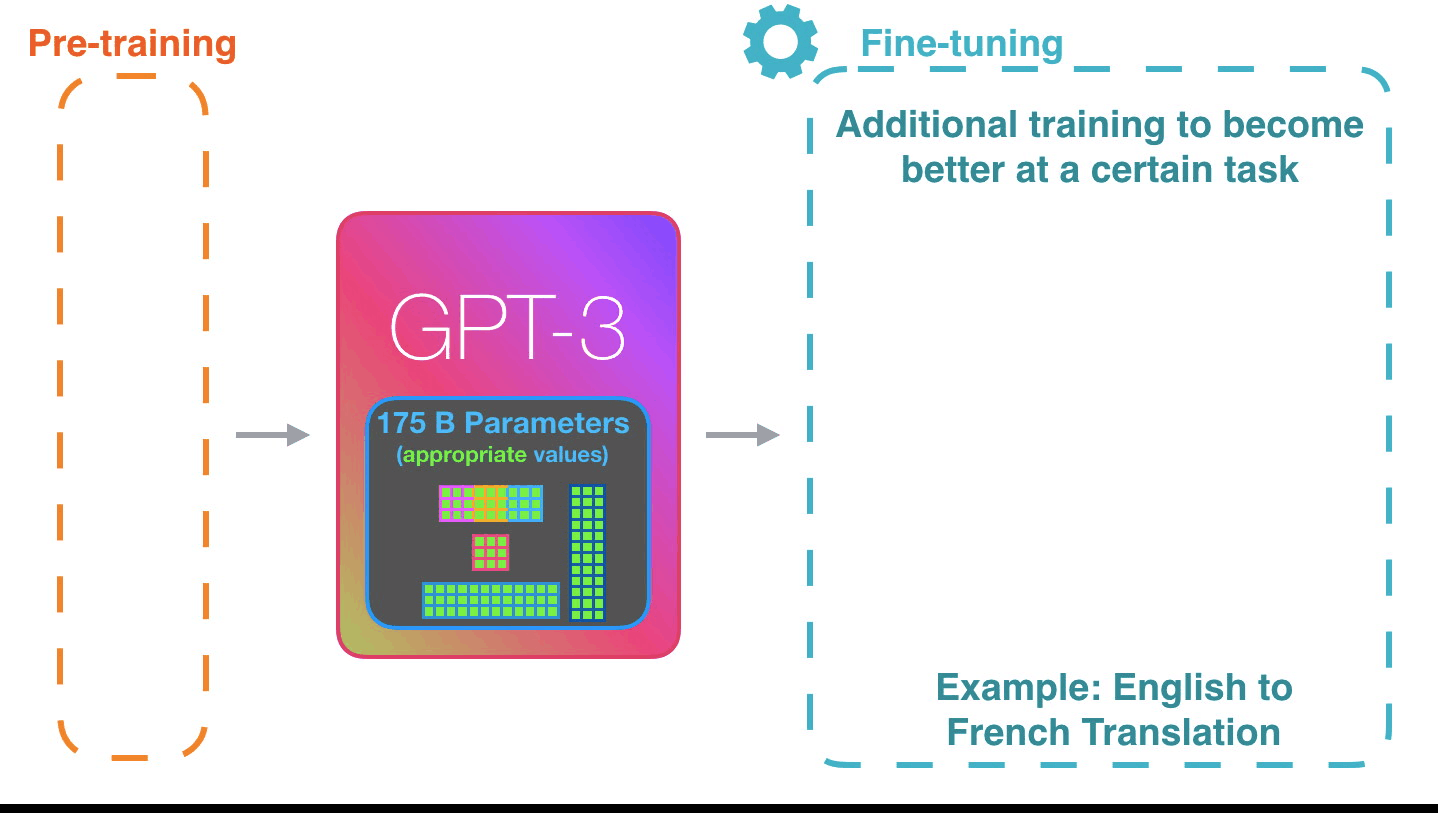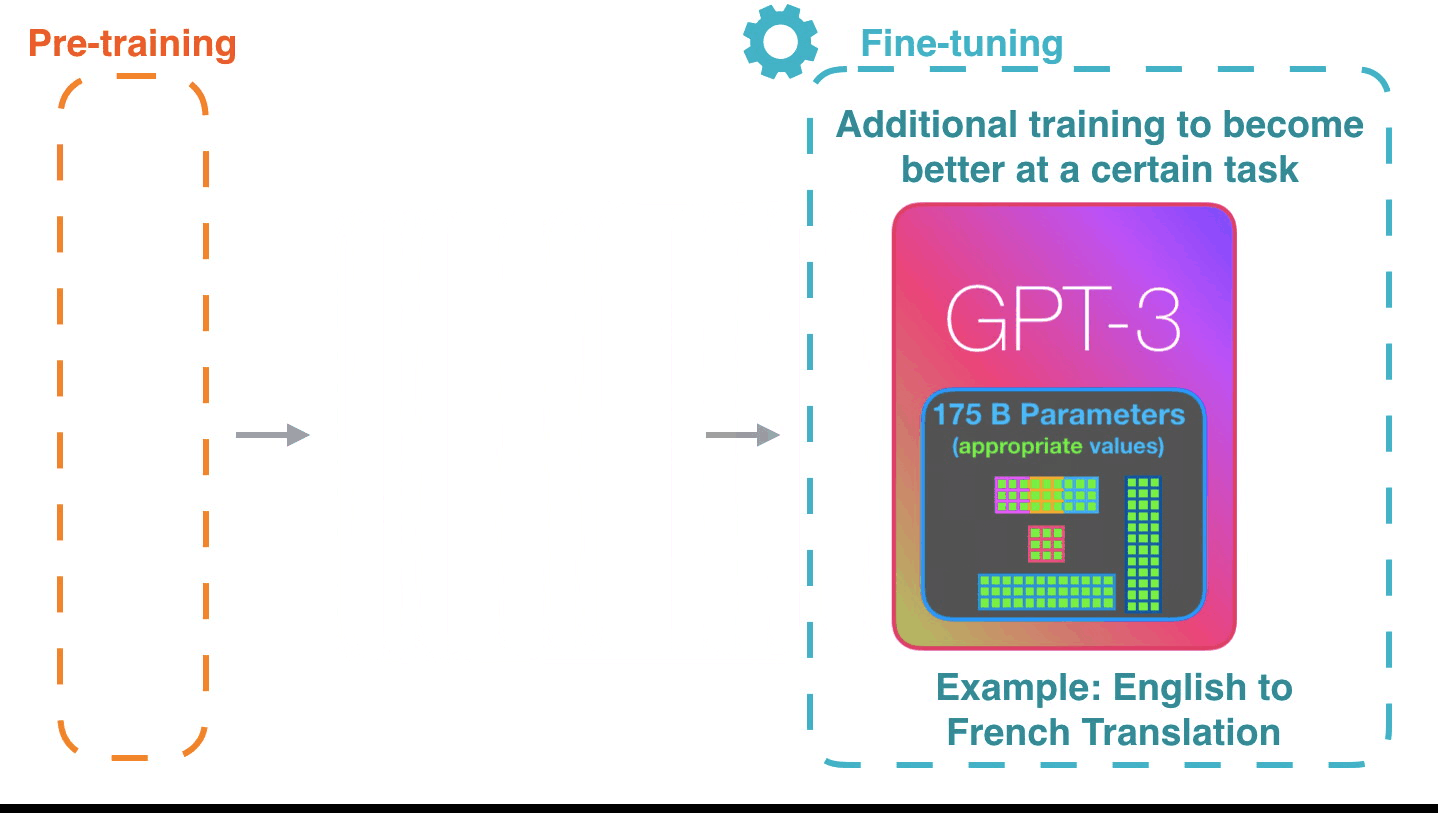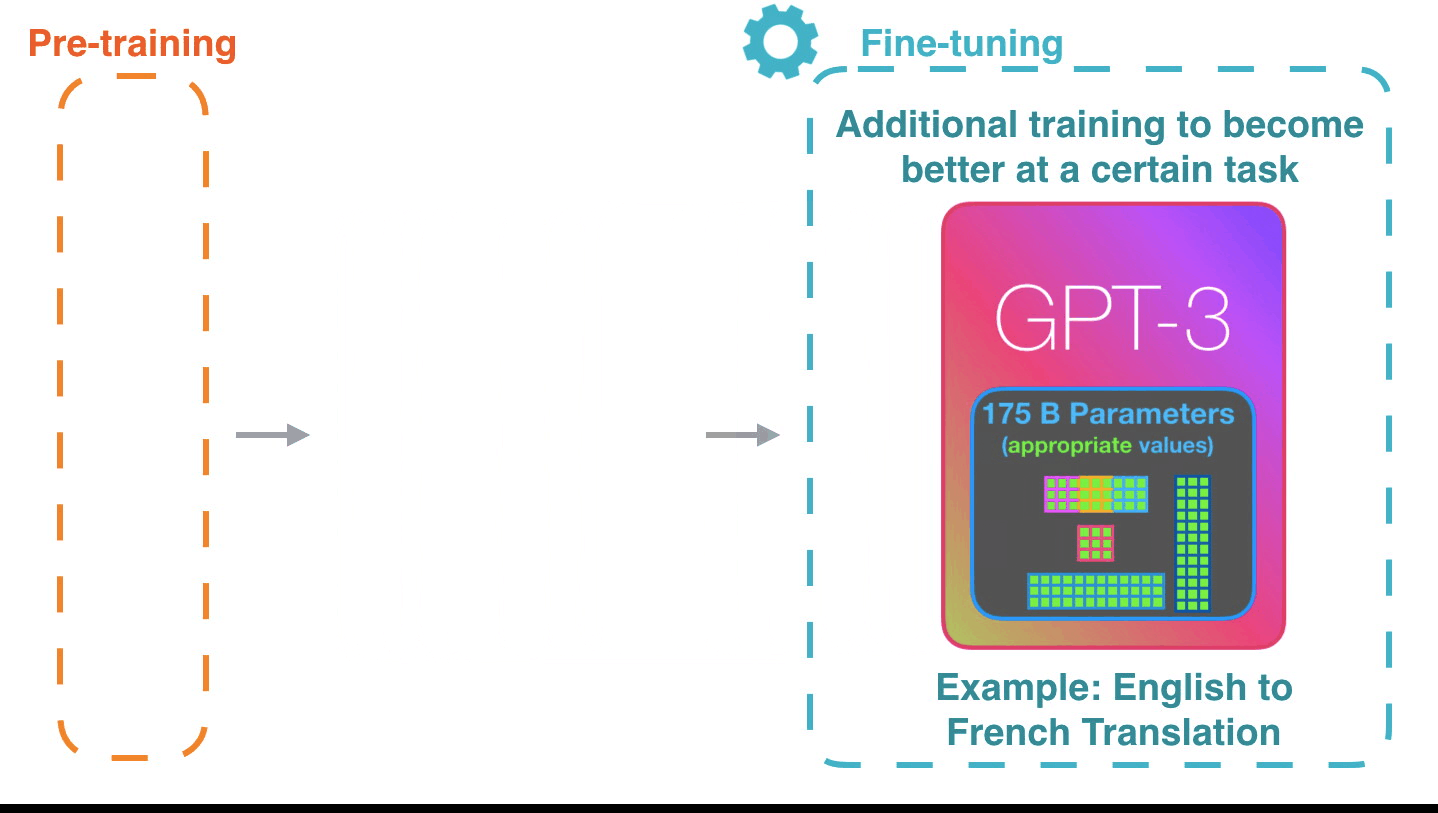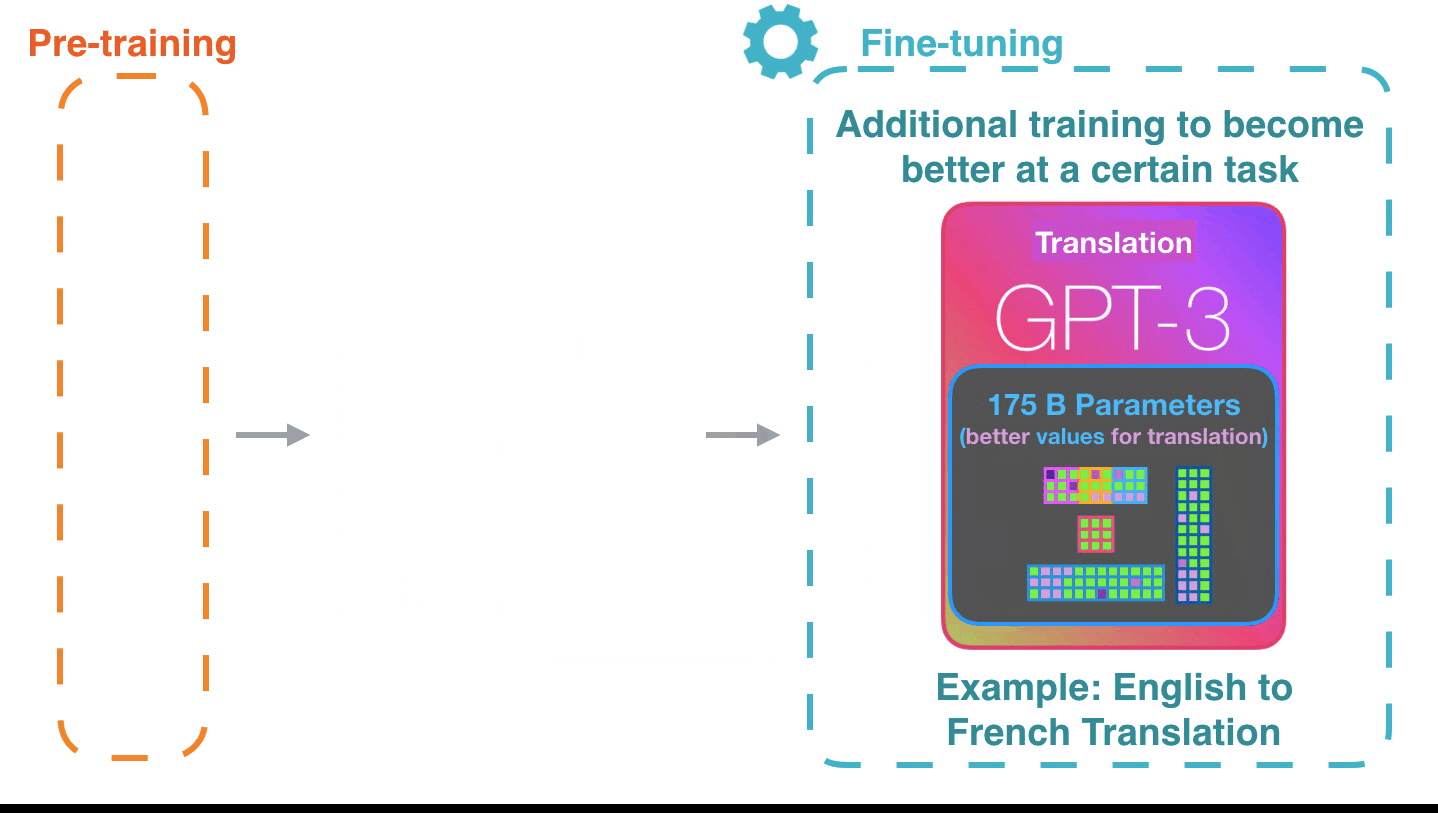
Assistant Models
saforem2/wordplay 🎮💬
- Fork of Andrej Karpathy’s
nanoGPT

saforem2/wordplay 🎮💬


Install
python3 -m pip install "git+https://github.com/saforem2/wordplay.git"
python3 -c 'import wordplay; print(wordplay.__file__)'
# ./wordplay/src/wordplay/__init__.py
Dependencies
-
transformers for transformers (to load GPT-2 checkpoints)
-
datasets for datasets (if you want to use OpenWebText)
-
tiktoken for OpenAI’s fast BPE code
-
wandb for optional logging
-
tqdm for progress bars
Quick Start
-
We start with training a character-level GPT on the works of Shakespeare.
- Downloading the data (~ 1MB) file
- Convert raw text to one large stream of integers
This will create data/shakespeare_char/{train.bin, val.bin}.
Model model.py
class CausalSelfAttention(nn.Module):
def __init__(self, config: GPTModelConfig):
super().__init__()
assert config.n_embd % config.n_head == 0
# key, query, value projections for all heads, but in a batch
self.c_attn = nn.Linear(
config.n_embd,
3 * config.n_embd,
bias=config.bias
)
# output projection
self.c_proj = nn.Linear(
config.n_embd,
config.n_embd,
bias=config.bias
)
# regularization
self.attn_dropout = nn.Dropout(config.dropout)
self.resid_dropout = nn.Dropout(config.dropout)
self.n_head = config.n_head
self.n_embd = config.n_embd
self.dropout = config.dropout
# flash attention make GPU go brrrrr but support is only in
# PyTorch >= 2.0
self.flash = hasattr(
torch.nn.functional,
'scaled_dot_product_attention'
)
# if self.flash and RANK == 0:
# log.warning(
# f'Using torch.nn.functional.scaled_dot_product_attention'
# '(Flash Attn)'
# )
if not self.flash:
log.warning(
"WARNING: using slow attention."
"Flash Attention requires PyTorch >= 2.0"
)
# causal mask to ensure that attention is only applied to the left
# in the input sequence
self.register_buffer(
"bias",
torch.tril(
torch.ones(
config.block_size,
config.block_size
)
).view(1, 1, config.block_size, config.block_size)
)
def forward(self, x):
# batch size, sequence length, embedding dimensionality (n_embd)
B, T, C = x.size()
# calculate query, key, values for all heads in batch and move head
# forward to be the batch dim
q, k, v = self.c_attn(x).split(self.n_embd, dim=2)
# (B, nh, T, hs)
k = k.view(B, T, self.n_head, C // self.n_head).transpose(1, 2)
# (B, nh, T, hs)
q = q.view(B, T, self.n_head, C // self.n_head).transpose(1, 2)
# (B, nh, T, hs)
v = v.view(B, T, self.n_head, C // self.n_head).transpose(1, 2)
# causal self-attention; Self-attend:
# (B, nh, T, hs) x (B, nh, hs, T) -> (B, nh, T, T)
if self.flash:
# efficient attention using Flash Attention CUDA kernels
y = torch.nn.functional.scaled_dot_product_attention(
q,
k,
v,
attn_mask=None,
dropout_p=(self.dropout if self.training else 0),
is_causal=True
)
else:
# manual implementation of attention
att = (q @ k.transpose(-2, -1)) * (1.0 / math.sqrt(k.size(-1)))
att = att.masked_fill(
self.bias[:, :, :T, :T] == 0, # type:ignore
float('-inf')
)
att = F.softmax(att, dim=-1)
att = self.attn_dropout(att)
y = att @ v # (B, nh, T, T) x (B, nh, T, hs) -> (B, nh, T, hs)
# re-assemble all head outputs side by side
y = y.transpose(1, 2).contiguous().view(B, T, C)
# output projection
y = self.resid_dropout(self.c_proj(y))
return y
class LayerNorm(nn.Module):
"""
LayerNorm but with an optional bias.
(PyTorch doesn't support simply bias=False)
"""
def __init__(self, ndim, bias):
super().__init__()
self.weight = nn.Parameter(torch.ones(ndim))
self.bias = nn.Parameter(torch.zeros(ndim)) if bias else None
def forward(self, input):
return F.layer_norm(
input,
self.weight.shape,
self.weight,
self.bias,
1e-5
)
class MLP(nn.Module):
def __init__(
self,
config: GPTModelConfig,
activation: str = 'gelu',
):
super().__init__()
self.c_fc = nn.Linear(
config.n_embd,
4 * config.n_embd,
bias=config.bias
)
if activation.lower() in ACTIVATIONS:
self.act_fn = ACTIVATIONS[activation.lower()]
else:
try:
act_fn = getattr(nn, activation)
assert callable(act_fn)
self.act_fn = act_fn()
except Exception as exc:
log.error(f'{activation} not yet supported!')
raise exc
# self.gelu = nn.GELU()
self.c_proj = nn.Linear(
4 * config.n_embd,
config.n_embd,
bias=config.bias
)
self.dropout = nn.Dropout(config.dropout)
def forward(self, x):
x = self.c_fc(x)
# x = self.gelu(x)
x = self.act_fn(x)
x = self.c_proj(x)
x = self.dropout(x)
return x
class Block(nn.Module):
def __init__(self, config: GPTModelConfig):
super().__init__()
self.ln_1 = LayerNorm(config.n_embd, bias=config.bias)
self.attn = CausalSelfAttention(config)
self.ln_2 = LayerNorm(config.n_embd, bias=config.bias)
self.mlp = MLP(config)
def forward(self, x):
x = x + self.attn(self.ln_1(x))
x = x + self.mlp(self.ln_2(x))
return x
class GPT(nn.Module):
def __init__(self, config: GPTModelConfig):
super().__init__()
assert config.vocab_size is not None
assert config.block_size is not None
self.config = config
self.transformer = nn.ModuleDict(dict(
wte=nn.Embedding(config.vocab_size, config.n_embd),
wpe=nn.Embedding(config.block_size, config.n_embd),
drop=nn.Dropout(config.dropout),
h=nn.ModuleList([Block(config) for _ in range(config.n_layer)]),
ln_f=LayerNorm(config.n_embd, bias=config.bias),
))
self.lm_head = nn.Linear(config.n_embd, config.vocab_size, bias=False)
# with weight tying when using torch.compile() some warnings get
# generated: "UserWarning: functional_call was passed multiple values
# for tied weights. This behavior is deprecated and will be an error in
# future versions" not 100% sure what this is, so far seems to be
# harmless. TODO investigate
# https://paperswithcode.com/method/weight-tying
self.transformer.wte.weight = self.lm_head.weight # type:ignore
# init all weights
self.apply(self._init_weights)
# apply special scaled init to the residual projections, per GPT-2
for pn, p in self.named_parameters():
if pn.endswith('c_proj.weight'):
torch.nn.init.normal_(
p,
mean=0.0,
std=0.02/math.sqrt(2 * config.n_layer)
)
# report number of parameters
log.info("number of parameters: %.2fM" % (self.get_num_params()/1e6,))
def get_num_params(self, non_embedding=True):
"""
Return the number of parameters in the model.
For non-embedding count (default), the position embeddings get
subtracted.
The token embeddings would too, except due to the parameter sharing
these params are actually used as weights in the final layer, so we
include them.
"""
n_params = sum(p.numel() for p in self.parameters())
if non_embedding:
n_params -= self.transformer.wpe.weight.numel() # type:ignore
return n_params
def _init_weights(self, module):
if isinstance(module, nn.Linear):
torch.nn.init.normal_(module.weight, mean=0.0, std=0.02)
if module.bias is not None:
torch.nn.init.zeros_(module.bias)
elif isinstance(module, nn.Embedding):
torch.nn.init.normal_(module.weight, mean=0.0, std=0.02)
def forward(self, idx, targets=None):
device = idx.device
b, t = idx.size()
assert t <= self.config.block_size, (
f"Cannot forward sequence of length {t}, "
"block size is only {self.config.block_size}"
)
pos = torch.arange(
0,
t,
dtype=torch.long,
device=device
) # shape (t)
# forward the GPT model itself
# token embeddings of shape (b, t, n_embd)
tok_emb = self.transformer.wte(idx) # type:ignore
# position embeddings of shape (t, n_embd)
pos_emb = self.transformer.wpe(pos) # type:ignore
x = self.transformer.drop(tok_emb + pos_emb) # type:ignore
for block in self.transformer.h: # type:ignore
x = block(x)
x = self.transformer.ln_f(x) # type:ignore
if targets is not None:
# if we are given some desired targets also calculate the loss
logits = self.lm_head(x)
loss = F.cross_entropy(
logits.view(
-1,
logits.size(-1)
),
targets.view(-1),
ignore_index=-1
)
else:
# inference-time mini-optimization: only forward the lm_head on the
# very last position
# note: using list [-1] to preserve the time dim
logits = self.lm_head(x[:, [-1], :])
loss = None
return logits, loss
def crop_block_size(self, block_size):
# model surgery to decrease the block size if necessary e.g. we may
# load the GPT2 pretrained model checkpoint (block size 1024) but want
# to use a smaller block size for some smaller, simpler model
assert block_size <= self.config.block_size
self.config.block_size = block_size
self.transformer.wpe.weight = ( # type:ignore
nn.Parameter(
self.transformer.wpe.weight[:block_size] # type:ignore
)
)
for block in self.transformer.h: # type:ignore
if hasattr(block.attn, 'bias'):
block.attn.bias = (
block.attn.bias[:, :, :block_size, :block_size]
)
@classmethod
def from_pretrained(cls, model_type, override_args=None):
assert model_type in {'gpt2', 'gpt2-medium', 'gpt2-large', 'gpt2-xl'}
override_args = override_args or {} # default to empty dict
# only dropout can be overridden see more notes below
assert all(k == 'dropout' for k in override_args)
from transformers import GPT2LMHeadModel
log.info(f"loading weights from pretrained gpt: {model_type=}")
# n_layer, n_head and n_embd are determined from model_type
# gpt2: 124M params
# gpt2-medium: 350M params
# gpt2-large: 774M params
# gpt2-xl: 1558M params
config_args = {
# 'baby-llama2': dict(n_layer=16, n_head=16, n_embed=1024),
# 'llama2-7b': dict(n_layer=32, n_head=32, n_embd=4096),
'gpt2': dict(n_layer=12, n_head=12, n_embd=768),
'gpt2-medium': dict(n_layer=24, n_head=16, n_embd=1024),
'gpt2-large': dict(n_layer=36, n_head=20, n_embd=1280),
'gpt2-xl': dict(n_layer=48, n_head=25, n_embd=1600),
}[model_type]
# we can override the dropout rate, if desired
if 'dropout' in override_args:
log.info(f"overriding dropout rate to {override_args['dropout']}")
config_args['dropout'] = override_args['dropout']
# create a from-scratch initialized minGPT model
log.info("forcing vocab_size=50257, block_size=1024, bias=True")
config = GPTModelConfig(
**config_args,
block_size=1024, # always 1024 for GPT model checkpoints
vocab_size=50257, # always 50257 for GPT model checkpoints
bias=True, # always True for GPT model checkpoints
)
model = GPT(config)
sd = model.state_dict()
sd_keys = sd.keys()
sd_keys = [
k for k in sd_keys if not k.endswith('.attn.bias')
] # discard this mask / buffer, not a param
# init a huggingface/transformers model
model_hf = GPT2LMHeadModel.from_pretrained(model_type)
sd_hf = model_hf.state_dict()
# copy while ensuring all of the parameters are aligned and match in
# names and shapes
sd_keys_hf = sd_hf.keys()
sd_keys_hf = [
k for k in sd_keys_hf if not k.endswith('.attn.masked_bias')
] # ignore these, just a buffer
sd_keys_hf = [
k for k in sd_keys_hf if not k.endswith('.attn.bias')
] # same, just the mask (buffer)
transposed = [
'attn.c_attn.weight',
'attn.c_proj.weight',
'mlp.c_fc.weight',
'mlp.c_proj.weight'
]
# basically the openai checkpoints use a "Conv1D" module, but we only
# want to use a vanilla Linear this means that we have to transpose
# these weights when we import them
assert len(sd_keys_hf) == len(sd_keys), (
f"mismatched keys: {len(sd_keys_hf)} != {len(sd_keys)}"
)
for k in sd_keys_hf:
if any(k.endswith(w) for w in transposed):
# special treatment for the Conv1D weights we need to transpose
assert sd_hf[k].shape[::-1] == sd[k].shape
with torch.no_grad():
sd[k].copy_(sd_hf[k].t())
else:
# vanilla copy over the other parameters
assert sd_hf[k].shape == sd[k].shape
with torch.no_grad():
sd[k].copy_(sd_hf[k])
return model
def configure_optimizers(
self,
weight_decay,
learning_rate,
betas,
device_type
):
# start with all of the candidate parameters
# filter out those that do not require grad
# param_dict = {
# pn: p for pn, p in param_dict.items() if p.requires_grad
# }
param_dict = {
pn: p for pn, p in self.named_parameters() if p.requires_grad
}
# create optim groups. Any parameters that is 2D will be weight
# decayed, otherwise no. i.e. all weight tensors in matmuls +
# embeddings decay, all biases and layernorms don't.
decay_params = [p for _, p in param_dict.items() if p.dim() >= 2]
nodecay_params = [p for _, p in param_dict.items() if p.dim() < 2]
optim_groups = [
{'params': decay_params, 'weight_decay': weight_decay},
{'params': nodecay_params, 'weight_decay': 0.0}
]
num_decay_params = sum(p.numel() for p in decay_params)
num_nodecay_params = sum(p.numel() for p in nodecay_params)
log.info(
f"num decayed parameter tensors: {len(decay_params)}, "
f"with {num_decay_params:,} parameters"
)
log.info(
f"num non-decayed parameter tensors: {len(nodecay_params)}, "
f"with {num_nodecay_params:,} parameters"
)
# Create AdamW optimizer and use the fused version if it is available
fused_available = (
'fused' in inspect.signature(torch.optim.AdamW).parameters
)
use_fused = fused_available and device_type == 'cuda'
extra_args = dict(fused=True) if use_fused else {}
optimizer = torch.optim.AdamW(
optim_groups,
lr=learning_rate,
betas=betas,
**extra_args
)
log.info(f"using fused AdamW: {use_fused}")
return optimizer
def estimate_mfu(self, fwdbwd_per_iter, dt):
"""Estimate model flops utilization (MFU)
(in units of A100 bfloat16 peak FLOPS)
"""
# first estimate the number of flops we do per iteration.
# see PaLM paper Appendix B as ref: https://arxiv.org/abs/2204.02311
N = self.get_num_params()
cfg = self.config
L, H, Q, T = (
cfg.n_layer,
cfg.n_head,
cfg.n_embd//cfg.n_head,
cfg.block_size
)
flops_per_token = 6*N + 12*L*H*Q*T
flops_per_fwdbwd = flops_per_token * T
flops_per_iter = flops_per_fwdbwd * fwdbwd_per_iter
# express our flops throughput as ratio of A100 bfloat16 peak flops
flops_achieved = flops_per_iter * (1.0/dt) # per second
flops_promised = 312e12 # A100 GPU bfloat16 peak flops is 312 TFLOPS
return flops_achieved / flops_promised
@torch.no_grad()
def generate(self, idx, max_new_tokens, temperature=1.0, top_k=None):
"""
Take a conditioning sequence of indices idx (LongTensor of shape (b,t))
and complete the sequence max_new_tokens times, feeding the predictions
back into the model each time.
Most likely you'll want to make sure to be in model.eval() mode of
operation for this.
"""
for _ in range(max_new_tokens):
# if the sequence context is growing too long we must crop it at
# block_size
idx_cond = (
idx if idx.size(1) <= self.config.block_size
else idx[:, -self.config.block_size:]
)
# forward the model to get the logits for the index in the sequence
logits, _ = self(idx_cond)
# pluck the logits at the final step and scale by desired
# temperature
logits = logits[:, -1, :] / temperature
# optionally crop the logits to only the top k options
if top_k is not None:
v, _ = torch.topk(logits, min(top_k, logits.size(-1)))
logits[logits < v[:, [-1]]] = -float('Inf')
# apply softmax to convert logits to (normalized) probabilities
probs = F.softmax(logits, dim=-1)
# sample from the distribution
idx_next = torch.multinomial(probs, num_samples=1)
# append sampled index to the running sequence and continue
idx = torch.cat((idx, idx_next), dim=1)
return idx
Trainer trainer.py
def get_batch(self, split: str) -> tuple[torch.Tensor, torch.Tensor]:
# data = self.config.train_data if split == 'train'
# else self.config.val_data
data = self.config.data.data.get(split, None)
assert data is not None
ix = torch.randint(
len(data) - self.config.model.block_size,
(self.config.model.batch_size,)
)
block_size = self.config.model.block_size
x = torch.stack(
[
torch.from_numpy((data[i:i+block_size]).astype(np.int64))
for i in ix
]
)
y = torch.stack(
[
torch.from_numpy((data[i+1:i+1+block_size]).astype(np.int64))
for i in ix
]
)
if self.config.device_type == 'cuda':
x = x.pin_memory().to(self.config.device_type, non_blocking=True)
y = y.pin_memory().to(self.config.device_type, non_blocking=True)
else:
x = x.to(self.config.device_type)
y = y.to(self.config.device_type)
return x, y
def _forward_step(self, x: torch.Tensor, y: torch.Tensor) -> dict:
t0 = time.perf_counter()
with self.config.ctx:
logits, loss = self.model_engine(x, y)
return {
'logits': logits,
'loss': loss,
'dt': time.perf_counter() - t0
}
def _backward_step(
self,
loss: torch.Tensor,
propagate_grads: bool = False,
) -> float:
t0 = time.perf_counter()
if self.config.train.backend.lower() in ['ds', 'deepspeed']:
self.model_engine.backward(loss) # type:ignore
self.model_engine.step(loss) # type:ignore
else:
if self.grad_scaler is not None:
self.grad_scaler.scale(loss).backward() # type:ignore
if propagate_grads:
if self.config.optimizer.grad_clip != 0.0:
if self.grad_scaler is not None:
self.grad_scaler.unscale_(self.optimizer)
torch.nn.utils.clip_grad_norm_( # pyright: ignore
self.model_engine.parameters(),
self.config.optimizer.grad_clip
)
if self.grad_scaler is not None:
self.grad_scaler.step(self.optimizer)
self.grad_scaler.update()
self.optimizer.zero_grad(set_to_none=True)
return time.perf_counter() - t0
def train_step(
self,
x: torch.Tensor,
y: torch.Tensor,
) -> dict:
lr = (
self.get_lr(self.config.iter_num)
if self.config.optimizer.decay_lr
else self._lr
)
for param_group in self.optimizer.param_groups:
param_group['lr'] = lr
dtf = []
dtb = []
dt = []
loss = torch.tensor(0.0)
for micro_step in range(self._gas):
is_last_micro_step = (micro_step == self._gas - 1)
# NOTE: -----------------------------------------------------------
# In DDP training we only need to sync gradients at the last micro
# step. the official way to do this is with model.no_sync() context
# manager, but I really dislike that this bloats the code and
# forces us to repeat code looking at the source of that context
# manager, it just toggles this variable
# -----------------------------------------------------------------
if self.config.train.backend.lower() == 'ddp':
_ = (
self.model_engine.require_backward_grad_sync
if (is_last_micro_step and self.world_size > 1)
else None
)
fout = self._forward_step(x, y)
# immediately async prefetch next batch while model is doing the
# forward pass on the GPU
x, y = self.get_batch('train')
loss = fout['loss'] / self._gas
dtf.append(fout['dt'])
dtb_ = self._backward_step(
loss,
propagate_grads=is_last_micro_step
)
dtb.append(dtb_)
dt.append(dtf + dtb)
timers = {
'iter': self.config.iter_num,
'dt': np.array(dt),
'dt_tot': np.sum(dt),
'dt_avg': np.mean(dt),
'dtf': np.array(dtf),
'dtf_tot': np.sum(dtf),
'dtf_avg': np.mean(dtf),
'dtb': np.array(dtb),
'dtb_tot': np.sum(dtb),
'dtb_avg': np.mean(dtb)
}
metrics = {
'iter': self.config.iter_num,
'loss': loss,
'lr': lr,
}
self.config.iter_num += 1
return {
'metrics': metrics,
'timers': timers,
'x': x,
'y': y,
}
def save_ckpt(
self,
raw_model: Optional[torch.nn.Module | GPT] = None,
add_to_wandb: bool = False
):
if raw_model is not None:
model = raw_model # type:ignore
else:
model = self.model
assert model is not None
assert isinstance(model, torch.nn.Module)
# assert issubclass(GPT, torch.nn.Module)
ckpt = {
'model': model.state_dict(),
'optimizer': self.optimizer.state_dict(),
'model_args': asdict(self.config.model),
'iter_num': self.config.iter_num,
'best_val_loss': self.config.best_val_loss,
'config': asdict(self.config),
}
# assert (
# isinstance(model, GPT)
# and issubclass(GPT, torch.nn.Module)
# )
# assert raw_model is not None
ckptfile = Path(os.getcwd()).joinpath('ckpt.pt')
modelfile = Path(os.getcwd()).joinpath('model.pth')
log.info(f'Saving checkpoint to: {os.getcwd()}')
log.info(f'Saving model to: {modelfile}')
torch.save(model.state_dict(), modelfile.as_posix())
torch.save(ckpt, ckptfile.as_posix())
add_to_ckpts_file(Path(os.getcwd()))
if add_to_wandb and wandb.run is not None:
artifact = wandb.Artifact('model', type='model')
artifact.add_file(modelfile.as_posix())
wandb.run.log_artifact(artifact)
def estimate_loss(self):
out = {}
self.model.eval()
for split in self.config.data.data.keys():
losses = torch.zeros(self.config.train.eval_iters)
for k in range(self.config.train.eval_iters):
x, y = self.get_batch(split)
with self.config.ctx:
_, loss = self.model_engine(x, y)
losses[k] = loss.item()
out[split] = losses.mean()
self.model.train()
return out
Hands-on Tutorial
Links
-
Hannibal046/Awesome-LLM 
- Mooler0410/LLMsPracticalGuide
- Large Language Models (in 2023)
- The Illustrated Transformer
- Generative AI Exists because of the Transformer
- GPT in 60 Lines of Numpy
-
Better Language Models and their Implications
-
Progress / Artefacts / Outcomes from 🌸 Bloom BigScience
[!NOTE]
Acknowledgements
This research used resources of the Argonne Leadership Computing Facility,
which is a DOE Office of Science User Facility supported under Contract DE-AC02-06CH11357.
References
Yao, Shunyu, Dian Yu, Jeffrey Zhao, Izhak Shafran, Thomas L. Griffiths, Yuan Cao, and Karthik Narasimhan. 2023. “Tree of Thoughts: Deliberate Problem Solving with Large Language Models.” https://arxiv.org/abs/2305.10601.
Citation
For attribution, please cite this work as:
Foreman, Sam. 2024. “Creating Small(-Ish) LLMs.” February
13. https://saforem2.github.io/LLM-tutorial.
window.document.addEventListener("DOMContentLoaded", function (event) {
const toggleBodyColorMode = (bsSheetEl) => {
const mode = bsSheetEl.getAttribute("data-mode");
const bodyEl = window.document.querySelector("body");
if (mode === "dark") {
bodyEl.classList.add("quarto-dark");
bodyEl.classList.remove("quarto-light");
} else {
bodyEl.classList.add("quarto-light");
bodyEl.classList.remove("quarto-dark");
}
}
const toggleBodyColorPrimary = () => {
const bsSheetEl = window.document.querySelector("link#quarto-bootstrap");
if (bsSheetEl) {
toggleBodyColorMode(bsSheetEl);
}
}
toggleBodyColorPrimary();
const disableStylesheet = (stylesheets) => {
for (let i=0; i < stylesheets.length; i++) {
const stylesheet = stylesheets[i];
stylesheet.rel = 'prefetch';
}
}
const enableStylesheet = (stylesheets) => {
for (let i=0; i < stylesheets.length; i++) {
const stylesheet = stylesheets[i];
stylesheet.rel = 'stylesheet';
}
}
const manageTransitions = (selector, allowTransitions) => {
const els = window.document.querySelectorAll(selector);
for (let i=0; i < els.length; i++) {
const el = els[i];
if (allowTransitions) {
el.classList.remove('notransition');
} else {
el.classList.add('notransition');
}
}
}
const toggleGiscusIfUsed = (isAlternate, darkModeDefault) => {
const baseTheme = document.querySelector('#giscus-base-theme')?.value ?? 'light';
const alternateTheme = document.querySelector('#giscus-alt-theme')?.value ?? 'dark';
let newTheme = '';
if(darkModeDefault) {
newTheme = isAlternate ? baseTheme : alternateTheme;
} else {
newTheme = isAlternate ? alternateTheme : baseTheme;
}
const changeGiscusTheme = () => {
// From: https://github.com/giscus/giscus/issues/336
const sendMessage = (message) => {
const iframe = document.querySelector('iframe.giscus-frame');
if (!iframe) return;
iframe.contentWindow.postMessage({ giscus: message }, 'https://giscus.app');
}
sendMessage({
setConfig: {
theme: newTheme
}
});
}
const isGiscussLoaded = window.document.querySelector('iframe.giscus-frame') !== null;
if (isGiscussLoaded) {
changeGiscusTheme();
}
}
const toggleColorMode = (alternate) => {
// Switch the stylesheets
const alternateStylesheets = window.document.querySelectorAll('link.quarto-color-scheme.quarto-color-alternate');
manageTransitions('#quarto-margin-sidebar .nav-link', false);
if (alternate) {
enableStylesheet(alternateStylesheets);
for (const sheetNode of alternateStylesheets) {
if (sheetNode.id === "quarto-bootstrap") {
toggleBodyColorMode(sheetNode);
}
}
} else {
disableStylesheet(alternateStylesheets);
toggleBodyColorPrimary();
}
manageTransitions('#quarto-margin-sidebar .nav-link', true);
// Switch the toggles
const toggles = window.document.querySelectorAll('.quarto-color-scheme-toggle');
for (let i=0; i < toggles.length; i++) {
const toggle = toggles[i];
if (toggle) {
if (alternate) {
toggle.classList.add("alternate");
} else {
toggle.classList.remove("alternate");
}
}
}
// Hack to workaround the fact that safari doesn't
// properly recolor the scrollbar when toggling (#1455)
if (navigator.userAgent.indexOf('Safari') > 0 && navigator.userAgent.indexOf('Chrome') == -1) {
manageTransitions("body", false);
window.scrollTo(0, 1);
setTimeout(() => {
window.scrollTo(0, 0);
manageTransitions("body", true);
}, 40);
}
}
const isFileUrl = () => {
return window.location.protocol === 'file:';
}
const hasAlternateSentinel = () => {
let styleSentinel = getColorSchemeSentinel();
if (styleSentinel !== null) {
return styleSentinel === "alternate";
} else {
return false;
}
}
const setStyleSentinel = (alternate) => {
const value = alternate ? "alternate" : "default";
if (!isFileUrl()) {
window.localStorage.setItem("quarto-color-scheme", value);
} else {
localAlternateSentinel = value;
}
}
const getColorSchemeSentinel = () => {
if (!isFileUrl()) {
const storageValue = window.localStorage.getItem("quarto-color-scheme");
return storageValue != null ? storageValue : localAlternateSentinel;
} else {
return localAlternateSentinel;
}
}
const darkModeDefault = false;
let localAlternateSentinel = darkModeDefault ? 'alternate' : 'default';
// Dark / light mode switch
window.quartoToggleColorScheme = () => {
// Read the current dark / light value
let toAlternate = !hasAlternateSentinel();
toggleColorMode(toAlternate);
setStyleSentinel(toAlternate);
toggleGiscusIfUsed(toAlternate, darkModeDefault);
};
// Ensure there is a toggle, if there isn't float one in the top right
if (window.document.querySelector('.quarto-color-scheme-toggle') === null) {
const a = window.document.createElement('a');
a.classList.add('top-right');
a.classList.add('quarto-color-scheme-toggle');
a.href = "";
a.onclick = function() { try { window.quartoToggleColorScheme(); } catch {} return false; };
const i = window.document.createElement("i");
i.classList.add('bi');
a.appendChild(i);
window.document.body.appendChild(a);
}
// Switch to dark mode if need be
if (hasAlternateSentinel()) {
toggleColorMode(true);
} else {
toggleColorMode(false);
}
const icon = "";
const anchorJS = new window.AnchorJS();
anchorJS.options = {
placement: 'right',
icon: icon
};
anchorJS.add('.anchored');
const viewSource = window.document.getElementById('quarto-view-source') ||
window.document.getElementById('quarto-code-tools-source');
if (viewSource) {
const sourceUrl = viewSource.getAttribute("data-quarto-source-url");
viewSource.addEventListener("click", function(e) {
if (sourceUrl) {
// rstudio viewer pane
if (/\bcapabilities=\b/.test(window.location)) {
window.open(sourceUrl);
} else {
window.location.href = sourceUrl;
}
} else {
const modal = new bootstrap.Modal(document.getElementById('quarto-embedded-source-code-modal'));
modal.show();
}
return false;
});
}
function toggleCodeHandler(show) {
return function(e) {
const detailsSrc = window.document.querySelectorAll(".cell > details > .sourceCode");
for (let i=0; i<detailsSrc.length; i++) {
const details = detailsSrc[i].parentElement;
if (show) {
details.open = true;
} else {
details.removeAttribute("open");
}
}
const cellCodeDivs = window.document.querySelectorAll(".cell > .sourceCode");
const fromCls = show ? "hidden" : "unhidden";
const toCls = show ? "unhidden" : "hidden";
for (let i=0; i<cellCodeDivs.length; i++) {
const codeDiv = cellCodeDivs[i];
if (codeDiv.classList.contains(fromCls)) {
codeDiv.classList.remove(fromCls);
codeDiv.classList.add(toCls);
}
}
return false;
}
}
const hideAllCode = window.document.getElementById("quarto-hide-all-code");
if (hideAllCode) {
hideAllCode.addEventListener("click", toggleCodeHandler(false));
}
const showAllCode = window.document.getElementById("quarto-show-all-code");
if (showAllCode) {
showAllCode.addEventListener("click", toggleCodeHandler(true));
}
function tippyHover(el, contentFn, onTriggerFn, onUntriggerFn) {
const config = {
allowHTML: true,
maxWidth: 500,
delay: 100,
arrow: false,
appendTo: function(el) {
return el.parentElement;
},
interactive: true,
interactiveBorder: 10,
theme: 'quarto',
placement: 'bottom-start',
};
if (contentFn) {
config.content = contentFn;
}
if (onTriggerFn) {
config.onTrigger = onTriggerFn;
}
if (onUntriggerFn) {
config.onUntrigger = onUntriggerFn;
}
window.tippy(el, config);
}
const noterefs = window.document.querySelectorAll('a[role="doc-noteref"]');
for (var i=0; i<noterefs.length; i++) {
const ref = noterefs[i];
tippyHover(ref, function() {
// use id or data attribute instead here
let href = ref.getAttribute('data-footnote-href') || ref.getAttribute('href');
try { href = new URL(href).hash; } catch {}
const id = href.replace(/^#\/?/, "");
const note = window.document.getElementById(id);
return note.innerHTML;
});
}
const xrefs = window.document.querySelectorAll('a.quarto-xref');
const processXRef = (id, note) => {
// Strip column container classes
const stripColumnClz = (el) => {
el.classList.remove("page-full", "page-columns");
if (el.children) {
for (const child of el.children) {
stripColumnClz(child);
}
}
}
stripColumnClz(note)
if (id === null || id.startsWith('sec-')) {
// Special case sections, only their first couple elements
const container = document.createElement("div");
if (note.children && note.children.length > 2) {
container.appendChild(note.children[0].cloneNode(true));
for (let i = 1; i < note.children.length; i++) {
const child = note.children[i];
if (child.tagName === "P" && child.innerText === "") {
continue;
} else {
container.appendChild(child.cloneNode(true));
break;
}
}
if (window.Quarto?.typesetMath) {
window.Quarto.typesetMath(container);
}
return container.innerHTML
} else {
if (window.Quarto?.typesetMath) {
window.Quarto.typesetMath(note);
}
return note.innerHTML;
}
} else {
// Remove any anchor links if they are present
const anchorLink = note.querySelector('a.anchorjs-link');
if (anchorLink) {
anchorLink.remove();
}
if (window.Quarto?.typesetMath) {
window.Quarto.typesetMath(note);
}
// TODO in 1.5, we should make sure this works without a callout special case
if (note.classList.contains("callout")) {
return note.outerHTML;
} else {
return note.innerHTML;
}
}
}
for (var i=0; i<xrefs.length; i++) {
const xref = xrefs[i];
tippyHover(xref, undefined, function(instance) {
instance.disable();
let url = xref.getAttribute('href');
let hash = undefined;
if (url.startsWith('#')) {
hash = url;
} else {
try { hash = new URL(url).hash; } catch {}
}
if (hash) {
const id = hash.replace(/^#\/?/, "");
const note = window.document.getElementById(id);
if (note !== null) {
try {
const html = processXRef(id, note.cloneNode(true));
instance.setContent(html);
} finally {
instance.enable();
instance.show();
}
} else {
// See if we can fetch this
fetch(url.split('#')[0])
.then(res => res.text())
.then(html => {
const parser = new DOMParser();
const htmlDoc = parser.parseFromString(html, "text/html");
const note = htmlDoc.getElementById(id);
if (note !== null) {
const html = processXRef(id, note);
instance.setContent(html);
}
}).finally(() => {
instance.enable();
instance.show();
});
}
} else {
// See if we can fetch a full url (with no hash to target)
// This is a special case and we should probably do some content thinning / targeting
fetch(url)
.then(res => res.text())
.then(html => {
const parser = new DOMParser();
const htmlDoc = parser.parseFromString(html, "text/html");
const note = htmlDoc.querySelector('main.content');
if (note !== null) {
// This should only happen for chapter cross references
// (since there is no id in the URL)
// remove the first header
if (note.children.length > 0 && note.children[0].tagName === "HEADER") {
note.children[0].remove();
}
const html = processXRef(null, note);
instance.setContent(html);
}
}).finally(() => {
instance.enable();
instance.show();
});
}
}, function(instance) {
});
}
let selectedAnnoteEl;
const selectorForAnnotation = ( cell, annotation) => {
let cellAttr = 'data-code-cell="' + cell + '"';
let lineAttr = 'data-code-annotation="' + annotation + '"';
const selector = 'span[' + cellAttr + '][' + lineAttr + ']';
return selector;
}
const selectCodeLines = (annoteEl) => {
const doc = window.document;
const targetCell = annoteEl.getAttribute("data-target-cell");
const targetAnnotation = annoteEl.getAttribute("data-target-annotation");
const annoteSpan = window.document.querySelector(selectorForAnnotation(targetCell, targetAnnotation));
const lines = annoteSpan.getAttribute("data-code-lines").split(",");
const lineIds = lines.map((line) => {
return targetCell + "-" + line;
})
let top = null;
let height = null;
let parent = null;
if (lineIds.length > 0) {
//compute the position of the single el (top and bottom and make a div)
const el = window.document.getElementById(lineIds[0]);
top = el.offsetTop;
height = el.offsetHeight;
parent = el.parentElement.parentElement;
if (lineIds.length > 1) {
const lastEl = window.document.getElementById(lineIds[lineIds.length - 1]);
const bottom = lastEl.offsetTop + lastEl.offsetHeight;
height = bottom - top;
}
if (top !== null && height !== null && parent !== null) {
// cook up a div (if necessary) and position it
let div = window.document.getElementById("code-annotation-line-highlight");
if (div === null) {
div = window.document.createElement("div");
div.setAttribute("id", "code-annotation-line-highlight");
div.style.position = 'absolute';
parent.appendChild(div);
}
div.style.top = top - 2 + "px";
div.style.height = height + 4 + "px";
div.style.left = 0;
let gutterDiv = window.document.getElementById("code-annotation-line-highlight-gutter");
if (gutterDiv === null) {
gutterDiv = window.document.createElement("div");
gutterDiv.setAttribute("id", "code-annotation-line-highlight-gutter");
gutterDiv.style.position = 'absolute';
const codeCell = window.document.getElementById(targetCell);
const gutter = codeCell.querySelector('.code-annotation-gutter');
gutter.appendChild(gutterDiv);
}
gutterDiv.style.top = top - 2 + "px";
gutterDiv.style.height = height + 4 + "px";
}
selectedAnnoteEl = annoteEl;
}
};
const unselectCodeLines = () => {
const elementsIds = ["code-annotation-line-highlight", "code-annotation-line-highlight-gutter"];
elementsIds.forEach((elId) => {
const div = window.document.getElementById(elId);
if (div) {
div.remove();
}
});
selectedAnnoteEl = undefined;
};
// Handle positioning of the toggle
window.addEventListener(
"resize",
throttle(() => {
elRect = undefined;
if (selectedAnnoteEl) {
selectCodeLines(selectedAnnoteEl);
}
}, 10)
);
function throttle(fn, ms) {
let throttle = false;
let timer;
return (...args) => {
if(!throttle) { // first call gets through
fn.apply(this, args);
throttle = true;
} else { // all the others get throttled
if(timer) clearTimeout(timer); // cancel #2
timer = setTimeout(() => {
fn.apply(this, args);
timer = throttle = false;
}, ms);
}
};
}
// Attach click handler to the DT
const annoteDls = window.document.querySelectorAll('dt[data-target-cell]');
for (const annoteDlNode of annoteDls) {
annoteDlNode.addEventListener('click', (event) => {
const clickedEl = event.target;
if (clickedEl !== selectedAnnoteEl) {
unselectCodeLines();
const activeEl = window.document.querySelector('dt[data-target-cell].code-annotation-active');
if (activeEl) {
activeEl.classList.remove('code-annotation-active');
}
selectCodeLines(clickedEl);
clickedEl.classList.add('code-annotation-active');
} else {
// Unselect the line
unselectCodeLines();
clickedEl.classList.remove('code-annotation-active');
}
});
}
const findCites = (el) => {
const parentEl = el.parentElement;
if (parentEl) {
const cites = parentEl.dataset.cites;
if (cites) {
return {
el,
cites: cites.split(' ')
};
} else {
return findCites(el.parentElement)
}
} else {
return undefined;
}
};
var bibliorefs = window.document.querySelectorAll('a[role="doc-biblioref"]');
for (var i=0; i<bibliorefs.length; i++) {
const ref = bibliorefs[i];
const citeInfo = findCites(ref);
if (citeInfo) {
tippyHover(citeInfo.el, function() {
var popup = window.document.createElement('div');
citeInfo.cites.forEach(function(cite) {
var citeDiv = window.document.createElement('div');
citeDiv.classList.add('hanging-indent');
citeDiv.classList.add('csl-entry');
var biblioDiv = window.document.getElementById('ref-' + cite);
if (biblioDiv) {
citeDiv.innerHTML = biblioDiv.innerHTML;
}
popup.appendChild(citeDiv);
});
return popup.innerHTML;
});
}
}
});
