https://github.com/scikit-learn-contrib/denmune-clustering-algorithm
DenMune a clustering algorithm that can find clusters of arbitrary size, shapes and densities in two-dimensions. Higher dimensions are first reduced to 2-D using the t-sne. The algorithm relies on a single parameter K (the number of nearest neighbors). The results show the superiority of DenMune. Enjoy the simplicty but the power of DenMune.
https://github.com/scikit-learn-contrib/denmune-clustering-algorithm
clustering deep-learning machine-learning python
Last synced: 3 months ago
JSON representation
DenMune a clustering algorithm that can find clusters of arbitrary size, shapes and densities in two-dimensions. Higher dimensions are first reduced to 2-D using the t-sne. The algorithm relies on a single parameter K (the number of nearest neighbors). The results show the superiority of DenMune. Enjoy the simplicty but the power of DenMune.
- Host: GitHub
- URL: https://github.com/scikit-learn-contrib/denmune-clustering-algorithm
- Owner: scikit-learn-contrib
- License: bsd-3-clause
- Created: 2021-12-21T04:09:37.000Z (over 3 years ago)
- Default Branch: main
- Last Pushed: 2025-03-07T04:10:45.000Z (4 months ago)
- Last Synced: 2025-03-24T19:05:24.108Z (3 months ago)
- Topics: clustering, deep-learning, machine-learning, python
- Language: Jupyter Notebook
- Homepage: https://mybinder.org/v2/gh/scikit-learn-contrib/denmune-clustering-algorithm/HEAD
- Size: 74 MB
- Stars: 31
- Watchers: 3
- Forks: 9
- Open Issues: 0
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
README
# DenMune: A Density-Peak Clustering Algorithm
DenMune is a clustering algorithm that can find clusters of arbitrary size, shapes, and densities in two-dimensions. Higher dimensions are first reduced to 2D using t-SNE. The algorithm relies on a single parameter K (the number of nearest neighbors). The results show the superiority of the algorithm. Enjoy the simplicity but the power of DenMune.
## Listen to this amazing interview podcast
[](https://on.soundcloud.com/z7WeqJnHjDd26hD76)
*click image to listen (24 min)*
## Reproducibility & Test Drives
Now you can reproduce all the research experiments, and even share the results and collaborate with the algorithm using our capsule on CodeOcean. Each Capsule is a self-contained computational experiment with computing environment, code, data, version history, and results.
Also, you may use our repo2docker offered by mybinder.org, which encapsulates the algorithm and all required data in one virtual machine instance. All Jupyter notebooks examples found in this repository will be also available to you in action to practice in this respo2docer. Thanks mybinder.org, you made it possible!
| Test-drive | URL |
| ---------------------------------------- | ------------------------------------------------------------ |
| Reproduce our code capsule on Code Ocean | [](https://bit.ly/codeocean-capsule) |
| Use our test-drive on MyBinder | [](https://bit.ly/mybinder-repo2docker) |
## Scientific Work
| Paper & data | Journals | ResearchGate Stats |
| :----------------------------------------------------------: | :----------------------------------------------------------: | :----------------------------------------------------------: |
| [](https://bit.ly/denmune-research-paper)
[](https://bit.ly/mendeley-data) | [](https://www.scimagojr.com/journalsearch.php?q=24823&tip=sid&clean=0) [](https://www.scimagojr.com/journalsearch.php?q=21101060167&tip=sid&clean=0) | 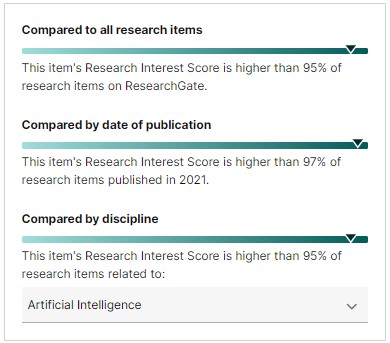 |
## Coding, Security & Maintenance
| Code Style | Installation | CI Workflow | Code Coverage | Code Scanning |
| ------------------------------------------------------------ | ------------------------------------------------------------ | ------------------------------------------------------------ | ------------------------------------------------------------ | ------------------------------------------------------------ |
|  | [](https://pypi.org/project/denmune/) | [](https://circleci.com/gh/egy1st/denmune-clustering-algorithm/tree/main) | [](https://codecov.io/gh/egy1st/denmune-clustering-algorithm) | [](https://github.com/adrinjalali/denmune-clustering-algorithm/actions/workflows/codeql.yml) |
## Tutorials
| Reproducible Capsule | Repo2Docker | Colab | Kaggle |
| ------------------------------------------------------------ | ------------------------------------------------------------ | ------------------------------------------------------------ | ------------------------------------------------------------ |
| [](https://bit.ly/codeocean-capsule) | [](https://bit.ly/mybinder-repo2docker) | [](#colab) | [](#kaggle) |
## Downloads Stats
| Download/Week | Download/Month | Total Downloads |
| ------------------------------------------------------------ | ------------------------------------------------------------ | ------------------------------------------------------------ |
| [](https://pepy.tech/project/denmune) | [](https://pepy.tech/project/denmune) | [](https://pepy.tech/project/denmune) |
## Based on the paper
| Paper |
|-
| Mohamed Abbas, Adel El-Zoghabi, Amin Shoukry,
| *DenMune: Density peak based clustering using mutual nearest neighbors*
| In: Journal of Pattern Recognition, Elsevier,
| volume 109, number 107589, January 2021
| DOI: https://doi.org/10.1016/j.patcog.2020.107589
## Documentation:
- [](https://denmune.readthedocs.io/en/latest/?badge=latest)
- [](https://denmune-docs.vercel.app)
## Watch it in action
This 30 seconds will tell you how a density-based algorithm, DenMune propagates:
[](https://colab.research.google.com/drive/1o-tP3uvDGjxBOGYkir1lnbr74sZ06e0U?usp=sharing)
[]()
## Still interested?
Watch this ***10-min*** illustrative video on:
- [](https://player.vimeo.com/video/827209757)
- [](https://www.youtube.com/watch?v=o77raaasuOM)
## When less means more
Most classic clustering algorithms fail to detect complex clusters where clusters are of different sizes, shapes, density, and exist in noisy data. Recently, a density-based algorithm named DenMune showed great ability in detecting complex shapes even in noisy data. it can detect a number of clusters automatically, detect both pre-identified-noise and post-identified-noise automatically, and remove them.
It can achieve an accuracy reach 100% in some classic pattern problems, achieve 97% in the MNIST dataset. A great advantage of this algorithm is being a single-parameter algorithm. All you need is to set a number of k-nearest neighbors and the algorithm will care about the rest. Being Non-sensitive to changes in k, make it robust and stable.
Keep in mind, that the algorithm reduces any N-D dataset to only a 2-D dataset initially, so it is a good benefit of this algorithm is always to plot your data and explore it which makes this algorithm a good candidate for data exploration. Finally, the algorithm comes with a neat package for visualizing data, validating it, and analyzing the whole clustering process.
## How to install DenMune
Simply install DenMune clustering algorithm using pip command from the official Python repository
[](https://pypi.org/project/denmune/)
From the shell run the command
```shell
pip install denmune
```
From Jupyter notebook cell run the command
```ipython3
!pip install denmune
```
## How to use DenMune
Once DenMune is installed, you just need to import it
```python
from denmune import DenMune
```
*Please note that first denmune (the package) in small letters, while the other one(the class itself) has D and M in capital case.*
## Read data
There are four possible cases of data:
- only train data without labels
- only labeled train data
- labeled train data in addition to test data without labels
- labeled train data in addition to labeled test data
```python
#=============================================
# First scenario: train data without labels
# ============================================
data_path = 'datasets/denmune/chameleon/'
dataset = "t7.10k.csv"
data_file = data_path + dataset
# train data without labels
X_train = pd.read_csv(data_file, sep=',', header=None)
knn = 39 # k-nearest neighbor, the only parameter required by the algorithm
dm = DenMune(train_data=X_train, k_nearest=knn)
labels, validity = dm.fit_predict(show_analyzer=False, show_noise=True)
```
This is an intuitive dataset which has no groundtruth provided

```python
#=============================================
# Second scenario: train data with labels
# ============================================
data_path = 'datasets/denmune/shapes/'
dataset = "aggregation.csv"
data_file = data_path + dataset
# train data with labels
X_train = pd.read_csv(data_file, sep=',', header=None)
y_train = X_train.iloc[:, -1]
X_train = X_train.drop(X_train.columns[-1], axis=1)
knn = 6 # k-nearest neighbor, the only parameter required by the algorithm
dm = DenMune(train_data=X_train, train_truth= y_train, k_nearest=knn)
labels, validity = dm.fit_predict(show_analyzer=False, show_noise=True)
```
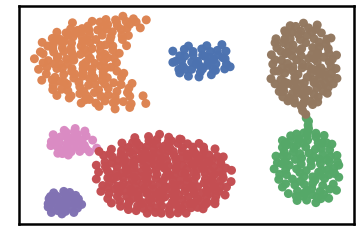
Datset groundtruth
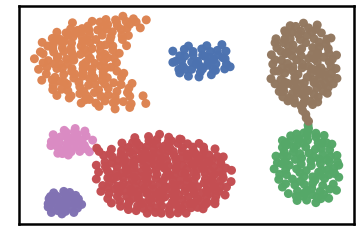
Dataset as detected by DenMune at k=6
```python
#=================================================================
# Third scenario: train data with labels in addition to test data
# ===============================================================
data_path = 'datasets/denmune/pendigits/'
file_2d = data_path + 'pendigits-2d.csv'
# train data with labels
X_train = pd.read_csv(data_path + 'train.csv', sep=',', header=None)
y_train = X_train.iloc[:, -1]
X_train = X_train.drop(X_train.columns[-1], axis=1)
# test data without labels
X_test = pd.read_csv(data_path + 'test.csv', sep=',', header=None)
X_test = X_test.drop(X_test.columns[-1], axis=1)
knn = 50 # k-nearest neighbor, the only parameter required by the algorithm
dm = DenMune(train_data=X_train, train_truth= y_train,
test_data= X_test,
k_nearest=knn)
labels, validity = dm.fit_predict(show_analyzer=True, show_noise=True)
```
dataset groundtruth
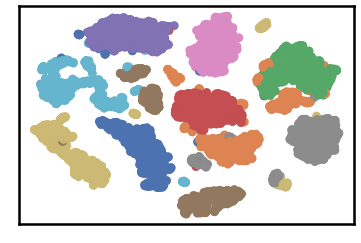
dataset as detected by DenMune at k=50
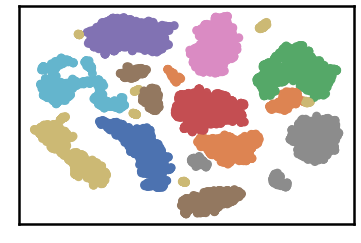
test data as predicted by DenMune on training the dataset at k=50
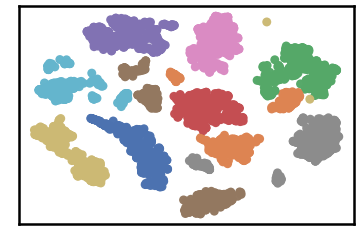
## Algorithm's Parameters
1. **Parameters used within the initialization of the DenMune class**
```python
def __init__ (self,
train_data=None, test_data=None,
train_truth=None, test_truth=None,
file_2d =None, k_nearest=1,
rgn_tsne=False, prop_step=0,
):
```
- train_data:
- data used for training the algorithm
- default: None. It should be provided by the use, otherwise an error will raise.
- train_truth:
- labels of training data
- default: None
- test_data:
- data used for testing the algorithm
- test_truth:
- labels of testing data
- default: None
- k_nearest:
- number of nearest neighbor
- default: 1. k-nearest neighbor should be at least 1.
- rgn_tsn:
- when set to True: It will regenerate the reduced 2-D version of the N-D dataset each time the algorithm run.
- when set to False: It will generate the reduced 2-D version of the N-D dataset first time only, then will reuse the saved exist file
- default: True
- file_2d: name (include location) of file used save/load the reduced 2-d version
- if empty: the algorithm will create temporary file named '_temp_2d'
- default: None
- prop_step:
- size of increment used in showing the clustering propagation.
- leave this parameter set to 0, the default value, unless you are willing intentionally to enter the propagation mode.
- default: 0
2. **Parameters used within the fit_predict function:**
```python
def fit_predict(self,
validate=True,
show_plots=True,
show_noise=True,
show_analyzer=True
):
```
- validate:
- validate data on/off according to five measures integrated with DenMune (Accuracy. F1-score, NMI index, AMI index, ARI index)
- default: True
- show_plots:
- show/hide plotting of data
- default: True
- show_noise:
- show/hide noise and outlier
- default: True
- show_analyzer:
- show/hide the analyzer
- default: True
## The Analyzer
The algorithm provide an exploratory tool called analyzer, once called it will provide you with in-depth analysis on how your clustering results perform.
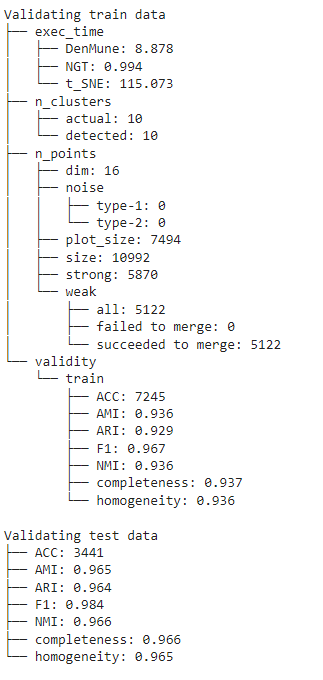
## Noise Detection
DenMune detects noise and outlier automatically, no need to any further work from your side.
- It plots pre-identified noise in black
- It plots post-identified noise in light grey
You can set show_noise parameter to False.
```python
# let us show noise
m = DenMune(train_data=X_train, k_nearest=knn)
labels, validity = dm.fit_predict(show_noise=True)
```
```python
# let us show clean data by removing noise
m = DenMune(train_data=X_train, k_nearest=knn)
labels, validity = dm.fit_predict(show_noise=False)
```
| noisy data | clean data |
| ------------------------------------------------------------ | ------------------------------------------------------------ |
|  |  |
## Validation
You can get your validation results using 3 methods
- by showing the Analyzer
- extract values from the validity returned list from fit_predict function
- extract values from the Analyzer dictionary
- There are five validity measures built-in the algorithm, which are:
- ACC, Accuracy
- F1 score
- NMI index (Normalized Mutual Information)
- AMI index (Adjusted Mutual Information)
- ARI index (Adjusted Rand Index)
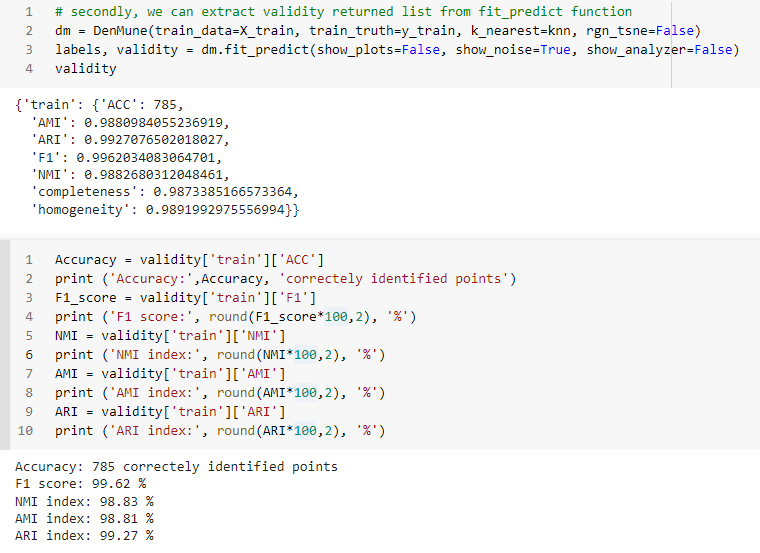
## K-nearest Evolution
The following chart shows the evolution of pre and post identified noise in correspondence to increase of number of knn. Also, detected number of clusters is analyzed in the same chart in relation with both types of identified noise.
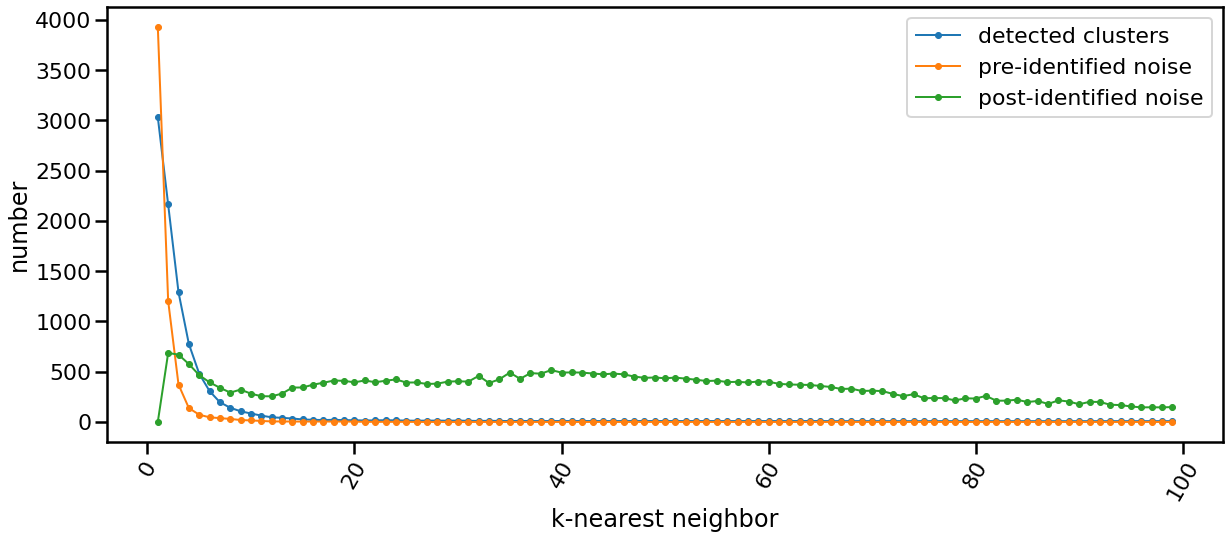
## The Scalability
| Data Size | Time |
| ----------------- | ---------------------- |
| data size: 5000 | time: 2.3139 seconds |
| data size: 10000 | time: 5.8752 seconds |
| data size: 15000 | time: 12.4535 seconds |
| data size: 20000 | time: 18.8466 seconds |
| data size: 25000 | time: 28.992 seconds |
| data size: 30000 | time: 39.3166 seconds |
| data size: 35000 | time: 39.4842 seconds |
| data size: 40000 | time: 63.7649 seconds |
| data size: 45000 | time: 73.6828 seconds |
| data size: 50000 | time: 86.9194 seconds |
| data size: 55000 | time: 90.1077 seconds |
| data size: 60000 | time: 125.0228 seconds |
| data size: 65000 | time: 149.1858 seconds |
| data size: 70000 | time: 177.4184 seconds |
| data size: 75000 | time: 204.0712 seconds |
| data size: 80000 | time: 220.502 seconds |
| data size: 85000 | time: 251.7625 seconds |
| data size: 100000 | time: 257.563 seconds |
|
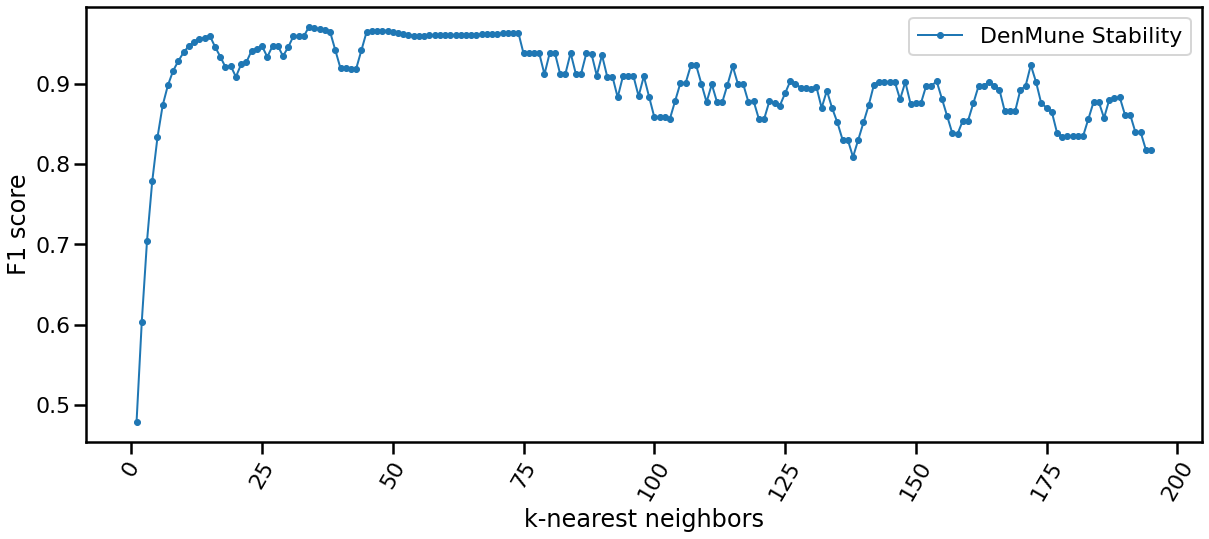
## The Stability
The algorithm is only single-parameter, even more it not sensitive to changes in that parameter, k. You may guess that from the following chart yourself. This is of great benefit for you as a data exploration analyst. You can simply explore the dataset using an arbitrary k. Being Non-sensitive to changes in k, make it robust and stable.
## Reveal the propagation
One of the top performing features in this algorithm is enabling you to watch how your clusters propagate to construct the final output clusters. Just use the parameter 'prop_step' as in the following example:
```python
dataset = "t7.10k" #
data_path = 'datasets/denmune/chameleon/'
# train file
data_file = data_path + dataset +'.csv'
X_train = pd.read_csv(data_file, sep=',', header=None)
from itertools import chain
# Denmune's Paramaters
knn = 39 # number of k-nearest neighbor, the only parameter required by the algorithm
# create list of differnt snapshots of the propagation
snapshots = chain(range(2,5), range(5,50,10), range(50, 100, 25), range(100,500,100), range(500,2000, 250), range(1000,5500, 500))
from IPython.display import clear_output
for snapshot in snapshots:
print ("itration", snapshot )
clear_output(wait=True)
dm = DenMune(train_data=X_train, k_nearest=knn, rgn_tsne=False, prop_step=snapshot)
labels, validity = dm.fit_predict(show_analyzer=False, show_noise=False)
```
## Interact with the algorithm
[](https://colab.research.google.com/drive/1EUROd6TRwxW3A_XD3KTxL8miL2ias4Ue?usp=sharing)
*click image to interact*
This notebook allows you interact with the algorithm in many aspects:
- you can choose which dataset to cluster (among 4 chameleon datasets)
- you can decide which number of k-nearest neighbor to use
- show noise on/off; thus you can invesetigate noise detected by the algorithm
- show analyzer on/off
## We love Jupyter Notebooks
Need to test examples one by one, then here other two options
- Use colab offered by google research to test each example individually.
- If you are a kaggler like me, then Kaggle, the best workspace where data scientist meet, should fit you to test the algorithm with great experience.
Here is a list of Google CoLab & Kaggle notebooks to practice the use of the algorithm interactively.
| Dataset | CoLab Notebook | Kaggle Notebook |
| ---------------------------- | ------------------------------------------------------------ | ------------------------------------------------------------ |
| How to use it? | [](https://bit.ly/colab-how-to-use) | [](https://bit.ly/kaggle-how-to-use) |
| Chameleon datasets | [](https://bit.ly/colab-chameleon) | [](https://bit.ly/kaggle-chameleon) |
| 2D Shape datasets | [](https://bit.ly/colab-2d-shapes) | [](https://bit.ly/kaggle-2d-shapes) |
| Clustering unlabeled data | [](https://bit.ly/colab-unlabeled-data) | [](https://bit.ly/kaggle-chameleon) |
| iris dataset | [](https://bit.ly/colab-iris-dataset) | [](https://bit.ly/kaggle-iris-dataset) |
| MNIST dataset | [](https://bit.ly/colab-mnist-dataset) | [](https://bit.ly/kaggle-score-97-mnist) |
| Scoring 97% on MNIST dataset | [](https://bit.ly/colab-score-97-mnist) | [](https://bit.ly/kaggle-score-97-mnist) |
| Noise detection | [](https://bit.ly/colab-noise-detection) | [](https://bit.ly/kaggle-noise-detection) |
| Validation | [](https://bit.ly/colab-how-to-validate) | [](https://bit.ly/kaggle-how-to-validate) |
| How does it propagate? | [](https://bit.ly/colab-how-propagate) | [](https://bit.ly/kaggle-how-propagate)
[](https://bit.ly/kaggle-how-propagate-2) |
| Snapshots of propagation | [](https://bit.ly/colab-propagation-shots) | [](https://bit.ly/kaggle-propagation-shots) |
| Scalability | [](https://bit.ly/colab-scalability) | [](https://bit.ly/kaggle-scalability) |
| Stability | [](https://bit.ly/colab-stability) | [](https://bit.ly/kaggle-stability) |
| k-nearest-evolution | [](https://bit.ly/colab-knn-evolution) | [](https://bit.ly/kaggle-knn-evolution) |
## Software Impact
Discover robust clustering without density cutoffs using this open-source Python library pyMune, implementing the parameter-free DenMune algorithm. PyMune identifies and expands cluster cores while removing noise. Fully scikit-learn compatible. pyMune (DenMune implementation) is a cutting-edge tool incorporating advanced techniques, robust performance, and effective propagation strategies. This positions it as the current state-of-the-art in its field, contributing to its high adoption and impact.
- After extensive research and rigorous validation, we are proud to release pyMune as an open-source tool on GitHub and PyPi for the benefit of the scientific community.
- With over 230,000 downloads already, pyMune has demonstrated its real-world impact and usefulness. We integrated it with [](https://bit.ly/codeocean-capsule) and [](https://bit.ly/mybinder-repo2docker) to further enhance reproducibility and reuse - encapsulating code, data, and outputs for turnkey sharing.
- It is part of a special issue of R-badged articles, https://www.sciencedirect.com/journal/software-impacts/special-issue/10XXN6LQ0J1
- it is part of Scikit-learn-contrib , https://github.com/scikit-learn-contrib

### Warning: Plagiarized Works
It has come to our attention that the following papers have plagiarized significant portions of the DenMune algorithm and research work:
1. **Paper 1:** "DEDIC: Density Estimation Clustering Method Using Directly Interconnected Cores" published in IEEE Access, doi: 10.1109/ACCESS.2022.3229582 Authors: Yisen Lin, Xinlun Zhang, Lei Liu, and Huichen Qu, reported at https://pubpeer.com/publications/AFC4E173A4FC0A2AD7E70DE688DDA5
2. **Paper 2:** "Research on stress curve clustering algorithm of Fiber Bragg grating sensor" published in Nature Scientific Reports, doi: 10.1038/s41598-023-39058-w Authors: Yisen Lin, Ye Wang, Huichen Qu & Yiwen Xiong, reported at https://pubpeer.com/publications/7AEF7D0F7505A8B8C130D142522741
We have conducted a thorough analysis and found extensive evidence of plagiarism in these papers, including:
- Verbatim copying of the core algorithm logic and steps from DenMune, with only superficial naming and implementation differences intended to obfuscate the similarity.
- Plagiarized background, related work, and technical details from the original DenMune paper, with minor paraphrasing and without proper attribution.
- Copying of mathematical formulations, concepts, and point classifications from DenMune.
- Reuse of experimental setup, datasets, and compared algorithms from DenMune without justification or acknowledgment.
- Fabricated experimental results, with values directly copied from DenMune's results and falsely claimed as their own.
- Lack of substantive analysis or discussion, further indicating that the experiments were likely not conducted.
Despite our efforts to address these concerns through proper channels, the publishers have decided to allow these plagiarized papers to remain published with only a correction acknowledging the issues, rather than retracting them or mandating a comprehensive correction.
We strongly condemn such academic misconduct and the potential enabling of plagiarism by reputable publishers. Researchers and practitioners should exercise caution when referring to or using the methods described in these plagiarized works.
For the original, properly cited implementation of the DenMune clustering algorithm, please refer to the official repository and resources provided here.
We remain committed to upholding academic integrity and ethical research practices, and we urge the scientific community to take a firm stance against plagiarism and misconduct in scholarly publications.
## How to cite
- How to cite ***The paper***
If you have used this codebase in a scientific publication and wish to cite it, please use the [Journal of Pattern Recognition article](https://www.sciencedirect.com/science/article/abs/pii/S0031320320303927):
```
Mohamed Abbas, Adel El-Zoghaby, Amin Shoukry, *DenMune: Density peak-based clustering using mutual nearest neighbors*
In: Journal of Pattern Recognition, Elsevier, volume 109, number 107589.
January 2021
```
```bib
@article{ABBAS2021107589,
title = {DenMune: Density peak-based clustering using mutual nearest neighbors},
journal = {Pattern Recognition},
volume = {109},
pages = {107589},
year = {2021},
issn = {0031-3203},
doi = {https://doi.org/10.1016/j.patcog.2020.107589},
url = {https://www.sciencedirect.com/science/article/pii/S0031320320303927},
author = {Mohamed Abbas and Adel El-Zoghabi and Amin Shoukry},
keywords = {Clustering, Mutual neighbors, Dimensionality reduction, Arbitrary shapes, Pattern recognition, Nearest neighbors, Density peak},
abstract = {Many clustering algorithms fail when clusters are of arbitrary shapes, of varying densities, or the data classes are unbalanced and close to each other, even in two dimensions. A novel clustering algorithm, “DenMune” is presented to meet this challenge. It is based on identifying dense regions using mutual nearest neighborhoods of size K, where K is the only parameter required from the user, besides obeying the mutual nearest neighbor consistency principle. The algorithm is stable for a wide range of values of K. Moreover, it is able to automatically detect and remove noise from the clustering process as well as detect the target clusters. It produces robust results on various low and high-dimensional datasets relative to several known state-of-the-art clustering algorithms.}
}
```
- How to cite ***The Software***
If you have used this codebase in a scientific publication and wish to cite it, please use the [Journal of Software Impacts article](https://www.sciencedirect.com/science/article/pii/S266596382300101X):
```
Abbas, M. A., El-Zoghabi, A., & Shoukry, A. (2023). PyMune: A Python package for complex clusters detection. Software Impacts, 17, 100564. https://doi.org/10.1016/j.simpa.2023.100564
```
```bib
@article{ABBAS2023100564,
title = {pyMune: A Python package for complex clusters detection},
journal = {Software Impacts},
volume = {17},
pages = {100564},
year = {2023},
issn = {2665-9638},
doi = {https://doi.org/10.1016/j.simpa.2023.100564},
url = {https://www.sciencedirect.com/science/article/pii/S266596382300101X},
author = {Mohamed Ali Abbas and Adel El-Zoghabi and Amin Shoukry},
keywords = {Machine learning, Pattern recognition, Dimensionality reduction, Mutual nearest neighbors, Nearest neighbors approximation, DenMune},
abstract = {We introduce pyMune, an open-source Python library for robust clustering of complex real-world datasets without density cutoff parameters. It implements DenMune (Abbas et al., 2021), a mutual nearest neighbor algorithm that uses dimensionality reduction and approximate nearest neighbor search to identify and expand cluster cores. Noise is removed with a mutual nearest-neighbor voting system. In addition to clustering, pyMune provides classification, visualization, and validation functionalities. It is fully compatible with scikit-learn and has been accepted into the scikit-learn-contrib repository. The code, documentation, and demos are available on GitHub, PyPi, and CodeOcean for easy use and reproducibility.}
}
```
## Licensing
The DenMune algorithm is 3-clause BSD licensed. Enjoy.
[](https://choosealicense.com/licenses/bsd-3-clause/)
## Task List
- [x] Update Github with the DenMune source code
- [x] create repo2docker repository
- [x] Create pip Package
- [x] create CoLab shared examples
- [x] create documentation
- [x] create Kaggle shared examples
- [x] PEP8 compliant
- [x] Continuous integration
- [x] scikit-learn compatible
- [x] creating unit tests (coverage: 100%)
- [x] generating API documentation
- [x] Create a reproducible capsule on code ocean
- [x] Submitting pyMune to Software Impacts (Published August 5 , 2023)
- [ ] create conda package (*postponed until NGT has conda installation*)