https://github.com/snap-stanford/Biomni
Biomni: a general-purpose biomedical AI agent
https://github.com/snap-stanford/Biomni
agent ai biomedicine
Last synced: 6 months ago
JSON representation
Biomni: a general-purpose biomedical AI agent
- Host: GitHub
- URL: https://github.com/snap-stanford/Biomni
- Owner: snap-stanford
- License: apache-2.0
- Created: 2025-03-19T21:06:53.000Z (10 months ago)
- Default Branch: main
- Last Pushed: 2025-07-09T18:31:56.000Z (6 months ago)
- Last Synced: 2025-07-10T01:05:43.506Z (6 months ago)
- Topics: agent, ai, biomedicine
- Language: Python
- Homepage: https://biomni.stanford.edu
- Size: 589 KB
- Stars: 581
- Watchers: 38
- Forks: 61
- Open Issues: 4
-
Metadata Files:
- Readme: README.md
Awesome Lists containing this project
- AiTreasureBox - snap-stanford/Biomni - 11-03_2301_0](https://img.shields.io/github/stars/snap-stanford/Biomni.svg)|Biomni: a general-purpose biomedical AI agent| (Repos)
- StarryDivineSky - snap-stanford/Biomni
- awesome-ai-agents - snap-stanford/Biomni - Biomni is a general-purpose biomedical AI agent that autonomously executes diverse research tasks across multiple biomedical subfields by integrating advanced LLM reasoning, retrieval-augmented planning, and code-based execution. (Autonomous Research & Content Generation / Prompt Libraries)
README

# Biomni: A General-Purpose Biomedical AI Agent
## Overview
Biomni is a general-purpose biomedical AI agent designed to autonomously execute a wide range of research tasks across diverse biomedical subfields. By integrating cutting-edge large language model (LLM) reasoning with retrieval-augmented planning and code-based execution, Biomni helps scientists dramatically enhance research productivity and generate testable hypotheses.
## Quick Start
### Installation
Our software environment is massive and we provide a single setup.sh script to setup.
Follow this [file](biomni_env/README.md) to setup the env first.
Then activate the environment E1:
```bash
conda activate biomni_e1
```
then install the latest biomni package:
```bash
pip install biomni --upgrade
```
Or install from the github source version.
Lastly, configure your API keys in bash profile `~/.bashrc`:
```bash
export ANTHROPIC_API_KEY="YOUR_API_KEY"
export OPENAI_API_KEY="YOUR_API_KEY" # optional if you just use Claude
```
### Basic Usage
Once inside the environment, you can start using Biomni:
```python
from biomni.agent import A1
# Initialize the agent with data path, Data lake will be automatically downloaded on first run (~11GB)
agent = A1(path='./data', llm='claude-sonnet-4-20250514')
# Execute biomedical tasks using natural language
agent.go("Plan a CRISPR screen to identify genes that regulate T cell exhaustion, generate 32 genes that maximize the perturbation effect.")
agent.go("Perform scRNA-seq annotation at [PATH] and generate meaningful hypothesis")
agent.go("Predict ADMET properties for this compound: CC(C)CC1=CC=C(C=C1)C(C)C(=O)O")
```
## 🤝 Contributing to Biomni
Biomni is an open-science initiative that thrives on community contributions. We welcome:
- **🔧 New Tools**: Specialized analysis functions and algorithms
- **📊 Datasets**: Curated biomedical data and knowledge bases
- **💻 Software**: Integration of existing biomedical software packages
- **📋 Benchmarks**: Evaluation datasets and performance metrics
- **📚 Misc**: Tutorials, examples, and use cases
- **🔧 Update existing tools**: many current tools are not optimized - fix and replacements are welcome!
Check out this **[Contributing Guide](CONTRIBUTION.md)** on how to contribute to the Biomni ecosystem.
If you have particular tool/database/software in mind that you want to add, you can also submit to [this form](https://forms.gle/nu2n1unzAYodTLVj6) and the biomni team will implement them.
## 🔬 Call for Contributors: Help Build Biomni-E2
Biomni-E1 only scratches the surface of what’s possible in the biomedical action space.
Now, we’re building **Biomni-E2** — a next-generation environment developed **with and for the community**.
We believe that by collaboratively defining and curating a shared library of standard biomedical actions, we can accelerate science for everyone.
**Join us in shaping the future of biomedical AI agent.**
- **Contributors with significant impact** (e.g., 10+ significant & integrated tool contributions or equivalent) will be **invited as co-authors** on our upcoming paper in a top-tier journal or conference.
- **All contributors** will be acknowledged in our publications.
- More contributor perks...
Let’s build it together.
## Tutorials and Examples
**[Biomni 101](./tutorials/biomni_101.ipynb)** - Basic concepts and first steps
More to come!
## 🌐 Web Interface
Experience Biomni through our no-code web interface at **[biomni.stanford.edu](https://biomni.stanford.edu)**.
[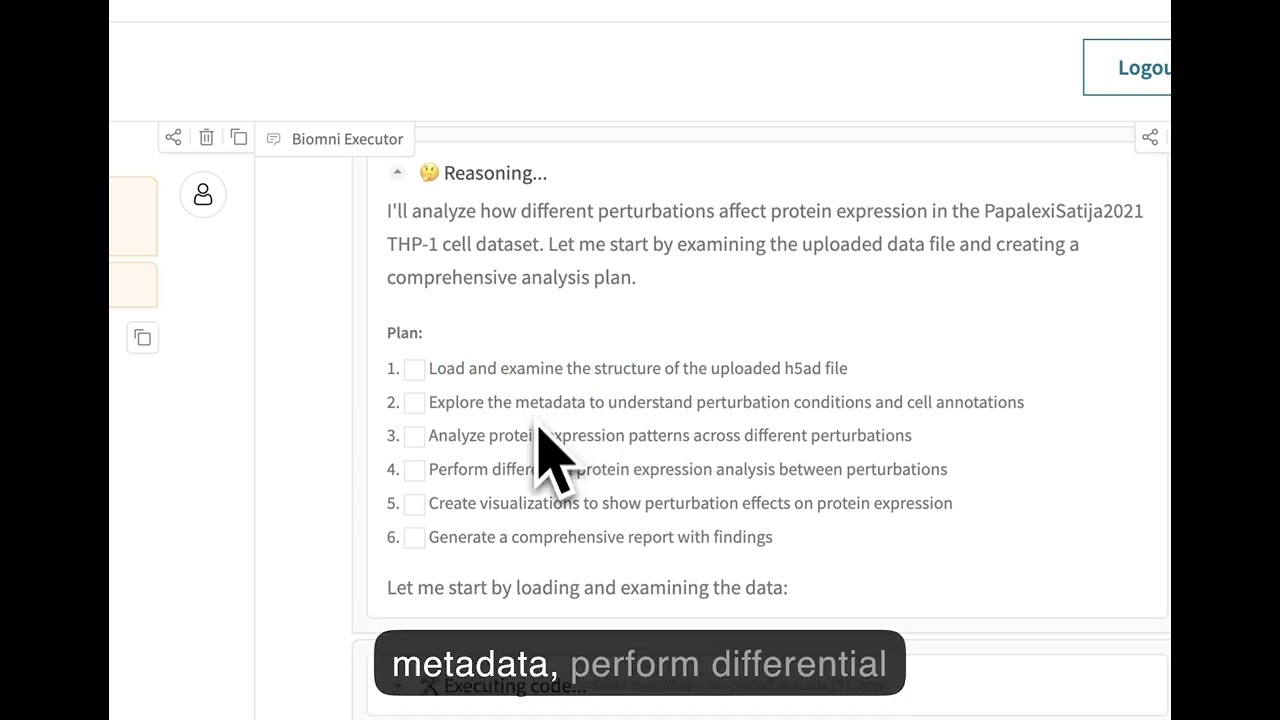](https://youtu.be/E0BRvl23hLs)
## Release schedule
- [ ] 8 Real-world research task benchmark/leaderboard release
- [ ] A tutorial on how to contribute to Biomni
- [ ] A tutorial on baseline agents
- [x] Biomni A1+E1 release
## Note
- This release was frozen as of April 15 2025, so it differs from the current web platform.
- Biomni itself is Apache 2.0-licensed, but certain integrated tools, databases, or software may carry more restrictive commercial licenses. Review each component carefully before any commercial use.
## Cite Us
```
@article{huang2025biomni,
title={Biomni: A General-Purpose Biomedical AI Agent},
author={Huang, Kexin and Zhang, Serena and Wang, Hanchen and Qu, Yuanhao and Lu, Yingzhou and Roohani, Yusuf and Li, Ryan and Qiu, Lin and Zhang, Junze and Di, Yin and others},
journal={bioRxiv},
pages={2025--05},
year={2025},
publisher={Cold Spring Harbor Laboratory}
}
```




