https://github.com/snf/keras-fractalnet
FractalNet implementation in Keras: Ultra-Deep Neural Networks without Residuals
https://github.com/snf/keras-fractalnet
Last synced: 7 months ago
JSON representation
FractalNet implementation in Keras: Ultra-Deep Neural Networks without Residuals
- Host: GitHub
- URL: https://github.com/snf/keras-fractalnet
- Owner: snf
- License: mit
- Created: 2016-06-23T19:07:57.000Z (over 9 years ago)
- Default Branch: master
- Last Pushed: 2017-09-17T17:11:23.000Z (about 8 years ago)
- Last Synced: 2024-10-31T17:39:12.575Z (about 1 year ago)
- Language: Python
- Homepage:
- Size: 2.6 MB
- Stars: 155
- Watchers: 9
- Forks: 38
- Open Issues: 0
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
- awesome-image-classification - unofficial-keras : https://github.com/snf/keras-fractalnet
- awesome-image-classification - unofficial-keras : https://github.com/snf/keras-fractalnet
README
# FractalNet implementation in Keras
## Information
I built this network as stated in the paper but fractals are done iterative instead of functional to avoid the extra complexity when merging the fractals.
The Join layers are built with a shared indicator sampled from a binomial distribution to indicate if global or local drop-path must be used.
When local drop-path is used, each Join layer samples it's own paths. But when global drop-path is used, all the join layers share the same tensor randomly sampled so one of the columns is globally selected.
## Notes
In the paper, they state that the last Join layer of each block is switched with the MaxPooling layer because of convenience. I don't do it and finish each block with a Join->MaxPooling but it should not affect the model.
Also it's not clear how and where the Dropout should be used. I found an implementation of the network [here](https://github.com/gustavla/fractalnet) by Larsson (one of the paper authors) and he adds it in each convolutional block (Convolution->Dropout->BatchNorm->ReLU). I implemented it the same way.
For testing the deepest column, the network is built with all the columns but the indicator for global drop-path is always set and the tensor with the paths is set to a constant array indicating which column is enabled.
## Model
Model graph image of FractalNet(c=3, b=5) generated by Keras: [link](https://raw.githubusercontent.com/snf/keras-fractalnet/master/doc/model.png)
## Experiments
This results are from the experiments with the code published here. The authors of the paper have not yet released a complete implementation of the network as of the publishing of this so I can't say what's different from theirs code. Also there is no kind of standardization, scaling or normalization across the dataset in these raw tests (which they may have used).
So far the results are promising when compared against Residual Networks. But I couldn't reproduce their deepest-column experiment.
The code here might have bugs too, if you find anything write me or submit a PR and I will rerun the tests.
Test error (%)
Method | C10 | C100
------ | --- | ----
ResNet (reported by [1]) | 13.63 | 44.76
ResNet Stochastic Depth (reported by [1]) | 11.66 | 37.80
FractalNet (paper w/SGD) | 10.18 | 35.34
FractalNet+dropout/drop-path (paper w/SGD) | 7.33 | 28.20
FractalNet+dropout/drop-path (this w/SGD) | 8.76 | 31.10
FractalNet+dropout/drop-path (this w/Adam) | 8.33 | 31.30
FractalNet+dropout/drop-path/deepest-column (paper w/SGD) | 7.27 | 29.05
FractalNet+dropout/drop-path/deepest-column (this w/SGD) | 12.53 | 43.07
FractalNet+dropout/drop-path/deepest-column (this w/Adam) | 12.28 | 41.32
[1] G. Huang, Y. Sun, Z. Liu, D. Sedra, and K. Weinberger. Deep networks with stochastic depth. arXiv preprint arXiv:1603.09382, 2016.
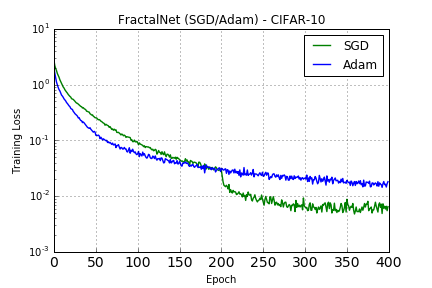
### CIFAR-10
Training as reported by the paper with SGD for 400 epochs starting with 0.02 learning rate and reducing it by 10x each time it reaches half of the remaining epochs (200, 300, 350, 375). Training with Adam is with default parameters.
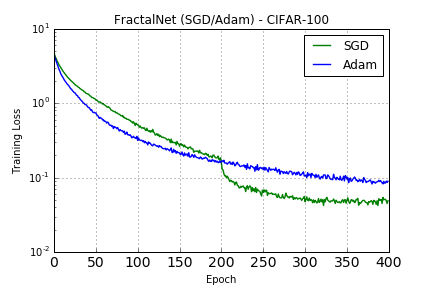
### CIFAR-100
Trained with SGD (as with CIFAR-10) and Adam with default parameters:
## Paper
arXiv: [FractalNet: Ultra-Deep Neural Networks without Residuals](https://arxiv.org/abs/1605.07648)
@article{larsson2016fractalnet,
title={FractalNet: Ultra-Deep Neural Networks without Residuals},
author={Larsson, Gustav and Maire, Michael and Shakhnarovich, Gregory},
journal={arXiv preprint arXiv:1605.07648},
year={2016}
}