Ecosyste.ms: Awesome
An open API service indexing awesome lists of open source software.
https://github.com/stat-ml/ncvis
Noise-Contrastive Visualization
https://github.com/stat-ml/ncvis
dimensionality-reduction machine-learning ncvis visualization
Last synced: 7 days ago
JSON representation
Noise-Contrastive Visualization
- Host: GitHub
- URL: https://github.com/stat-ml/ncvis
- Owner: stat-ml
- License: mit
- Created: 2019-07-01T14:08:25.000Z (over 5 years ago)
- Default Branch: master
- Last Pushed: 2023-11-25T17:57:22.000Z (12 months ago)
- Last Synced: 2024-04-24T15:42:49.912Z (7 months ago)
- Topics: dimensionality-reduction, machine-learning, ncvis, visualization
- Language: C++
- Size: 1.78 MB
- Stars: 51
- Watchers: 6
- Forks: 2
- Open Issues: 3
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
README
[](https://anaconda.org/conda-forge/ncvis)
[](https://pypi.python.org/pypi/ncvis/)
[](https://github.com/alartum/ncvis/blob/master/LICENSE)
[](https://anaconda.org/conda-forge/ncvis)
[](https://dev.azure.com/conda-forge/feedstock-builds/_build/latest?definitionId=8934&branchName=main)
[](https://anaconda.org/conda-forge/ncvis)
# ncvis
**NCVis** is an efficient solution for data visualization and dimensionality reduction. It uses [HNSW](https://github.com/nmslib/hnswlib) to quickly construct the nearest neighbors graph and a parallel (batched) approach to build its embedding. Efficient random sampling is achieved via [PCGRandom](https://github.com/imneme/pcg-cpp). Detailed application examples can be found [here](https://github.com/alartum/ncvis-examples).
# Why NCVis?
## It is Fast
We use preprocessed samples from the [News Headlines Of India dataset](https://www.kaggle.com/therohk/india-headlines-news-dataset) to perform the comparison. Test cases are generated by taking the first 1000, 2 · 1000, . . . , 2¹⁰ · 1000 samples from the dataset. Given the same amount of time **NCVis** allows to process more than double number of samples compared to other methods, visualizing **10⁶** points in only **6** minutes (12 × Intel® CoreTM i7-8700K CPU @
3.70GHz, 64 Gb RAM).

## It is Efficient
One can define efficiency as the ratio of the time to execute the task on a single processor to the time on multiple processors. Ideally, the efficiency should be equal to the num-
ber of threads. **NCVis** does not achieve this limit but signifi-
cantly outperforms other methods. We used 10000 samples from the [News Headlines Of India dataset](https://www.kaggle.com/therohk/india-headlines-news-dataset).

## It is Predictable
It is important that the proposed method has predictable behavior on simple datasets. We used the [Optical Recognition of Handwritten Digits Data Set](https://archive.ics.uci.edu/ml/datasets/optical+recognition+of+handwritten+digits) which comprised 5620 preprocessed handwritten digits and thus has a simple structure that is assumed to be revealed by visualization. **NCVis** shows the behavior consistent with classical methods like t-SNE while producing visualization up to the order of magnitude faster.
| t-SNE (29.5s) | FIt-SNE (17.4s) |
:-------------------------:|:-------------------------:
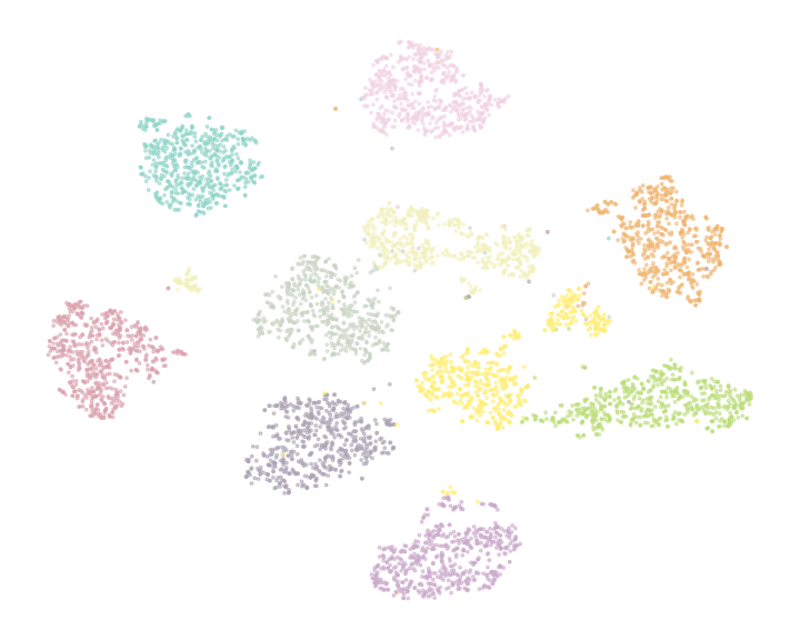 |
| 
| Multicore t-SNE (14.3s) | LargeVis (9.7s)|
:-------------------------:|:-------------------------:
 |
| 
| Umap (7.5s) | NCVis (0.9s)|
:-------------------------:|:-------------------------:
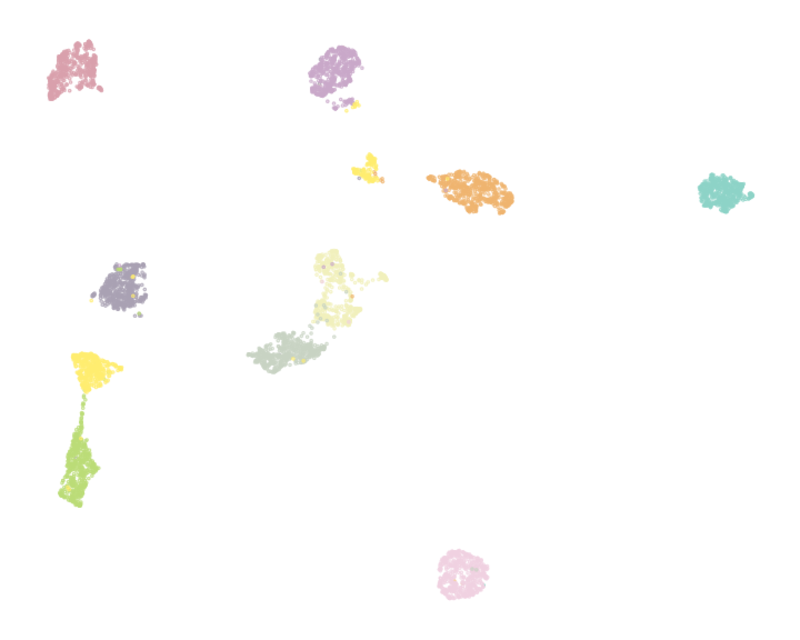 |
| 
# Using
```python
import ncvis
vis = ncvis.NCVis()
Y = vis.fit_transform(X)
```
More detailed examples can be found [here](https://github.com/alartum/ncvis-examples).
# Installation
## Conda [recommended]
You do not need to setup the environment if using *conda*, all dependencies are installed automatically. Using *conda-forge* channel is preferred, but using *alartum* channel is also possible in case of any issues with *conda-forge*.
```bash
$ conda install conda-forge::ncvis
```
or
```bash
$ conda install alartum::ncvis
```
## Pip [not recommended]
**Important**: be sure to have a compiler with *OpenMP* support. *GCC* has it by default, which is not the case with *clang*. You may need to install *llvm-openmp* library beforehand.
1. Install **numpy**, **cython** and **pybind11** packages (compile-time dependencies):
```bash
$ pip install numpy cython pybind11
```
2. Install **ncvis** package:
```bash
$ pip install ncvis
```
## From source [not recommended]
**Important**: be sure to have *OpenMP* available.
First of all, download the *pcg-cpp* and *hnswlib* libraries:
```bash
$ make libs
```
### Python Wrapper
If *conda* environment is used, it replaces library search paths. To prevent compilation errors, you either need to use compilers provided by *conda* or switch to *pip* and system compilers.
* Conda
```bash
$ conda install -c conda-forge cxx-compiler c-compiler conda-build numpy cython pybind11 scipy
$ conda-develop -bc .
```
* Pip
```bash
$ pip install numpy cython pybind11
$ make wrapper
```
You can then use *pytest* to run some basic checks
```bash
$ pytest -v recipe/test.py
```
### C++ Binary
* Release
```bash
$ make ncvis
```
* Debug
```bash
$ make debug
```
# Citation
The original paper can be found [here](https://dl.acm.org/doi/abs/10.1145/3366423.3380061). If you use **NCVis**, we kindly ask you to cite:
```
@inproceedings{10.1145/3366423.3380061,
author = {Artemenkov, Aleksandr and Panov, Maxim},
title = {NCVis: Noise Contrastive Approach for Scalable Visualization},
year = {2020},
isbn = {9781450370233},
publisher = {Association for Computing Machinery},
address = {New York, NY, USA},
url = {https://doi.org/10.1145/3366423.3380061},
doi = {10.1145/3366423.3380061},
booktitle = {Proceedings of The Web Conference 2020},
pages = {2941–2947},
numpages = {7},
keywords = {dimensionality reduction, noise contrastive estimation, embedding algorithms, visualization},
location = {Taipei, Taiwan},
series = {WWW ’20}
}
```