https://github.com/theislab/mapqc
MapQC - a metric for the evaluation of single-cell query-to-reference mappings
https://github.com/theislab/mapqc
atlas integration mapping query-to-reference-mapping single-cell transcriptomics
Last synced: 4 months ago
JSON representation
MapQC - a metric for the evaluation of single-cell query-to-reference mappings
- Host: GitHub
- URL: https://github.com/theislab/mapqc
- Owner: theislab
- License: mit
- Created: 2025-03-07T09:47:27.000Z (11 months ago)
- Default Branch: main
- Last Pushed: 2025-10-06T20:12:22.000Z (4 months ago)
- Last Synced: 2025-10-06T22:18:35.403Z (4 months ago)
- Topics: atlas, integration, mapping, query-to-reference-mapping, single-cell, transcriptomics
- Language: Python
- Homepage: https://mapqc.readthedocs.io/en/latest/
- Size: 31.8 MB
- Stars: 9
- Watchers: 0
- Forks: 1
- Open Issues: 2
-
Metadata Files:
- Readme: README.md
- Changelog: CHANGELOG.md
- Contributing: docs/contributing.md
- License: LICENSE
Awesome Lists containing this project
README
# mapQC
[![Tests][badge-tests]][tests]
[![Documentation][badge-docs]][documentation]
[badge-tests]: https://img.shields.io/github/actions/workflow/status/theislab/mapqc/test.yaml?branch=main
[badge-docs]: https://img.shields.io/readthedocs/mapqc
A metric for the evaluation of single-cell query-to-reference mappings
## Getting started
Please refer to the [documentation](https://mapqc.readthedocs.io/), in particular, the [API documentation](https://mapqc.readthedocs.io/en/latest/api/index.html) for detailed package documentation. For reproduction of the results in the paper, check out the [mapQC reproducibility repository](https://github.com/theislab/mapqc_reproducibility).
Below a few notes on how and when to use mapQC:
### What does mapQC do?
MapQC evaluates the quality of a query-to-reference mapping, and outputs a cell-level mapQC score for every query cell. MapQC scores higher than 2 indicate a large distance of the query cell to the reference. Given a healthy/control reference, we expect query controls to have low mapQC scores, and query case/disease cells to have higher mapQC scores in the case of case-specific cellular phenotypes. You can thus use mapQC scores to assess, in a quantitative manner, if your mapping was successful.
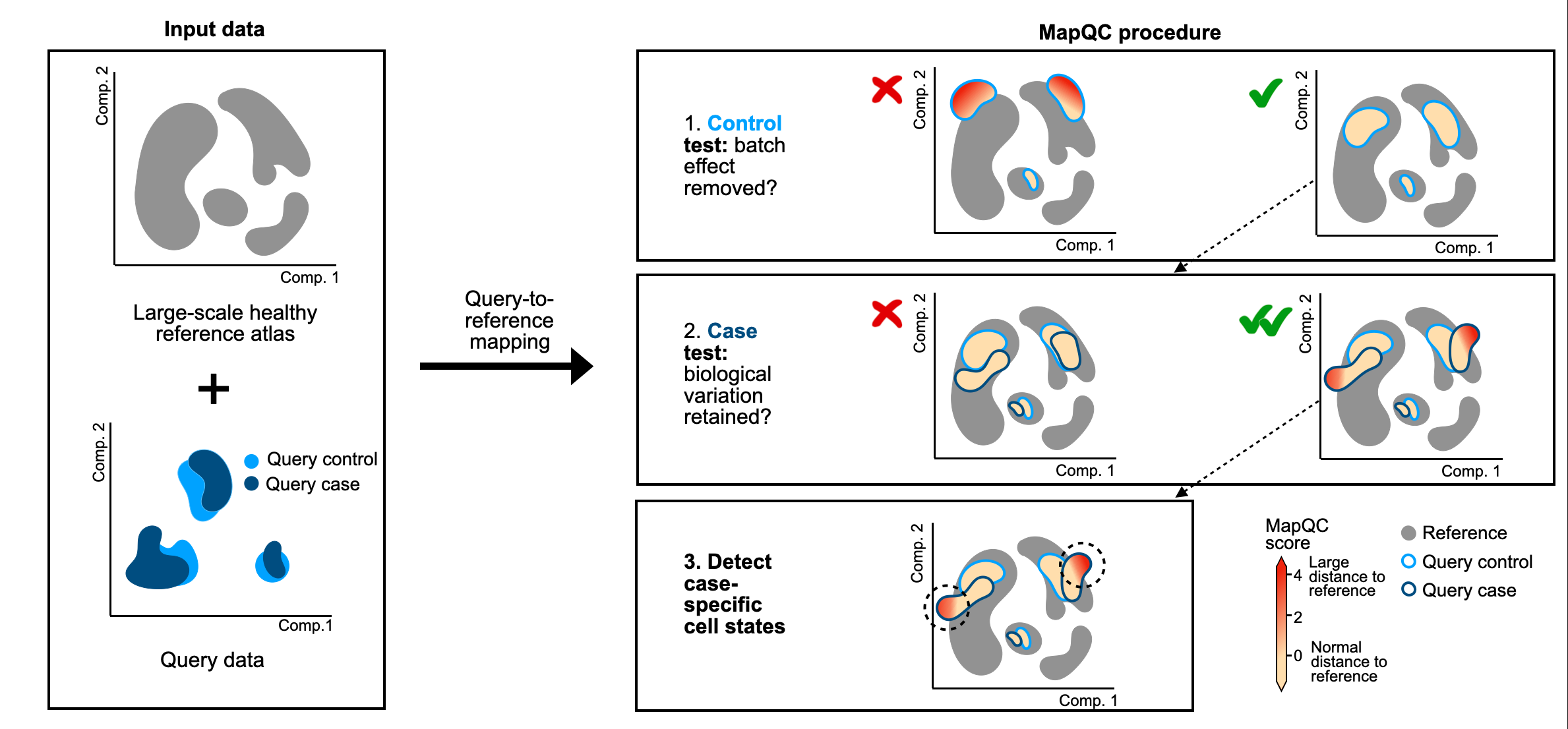
Overview of mapQC's workflow
### What are the data requirements for using mapQC?
In short, you need one AnnData object, including:
- A large scale reference, including only its healthy/control cells.
- A mapped query dataset, with healthy/control cells (must-have) and case/perturbed cells (if you have them).
- Metadata (query/reference status, study, sample, and optionally clustering and cell type annotations)
- A mapping-derived embedding including both the reference and the query
In the [quick-start tutorial notebook](./docs/notebooks/mapqc_quickstart.ipynb) we provide a more extensive description of the exact data requirements.
## Installation
You need to have Python 3.10 or newer installed on your system.
There are several alternative options to install mapQC:
1) Install the latest release of `mapqc` from [PyPI][]:
```bash
pip install mapqc
```
1. Install the latest development version:
```bash
pip install git+https://github.com/theislab/mapqc.git@main
```
## Release notes
See the [changelog][].
## Contact
I am happy to hear any comments, suggestions, or even bugs that you run into. I would like to make this package run as smoothly as possible! So for any of these, submit an issue on the mapQC GitHub page and I will be glad to help.
## Citation
[Sikkema et al.](https://www.biorxiv.org/content/10.1101/2025.05.23.655749v1), bioRxiv 2025.
[mambaforge]: https://github.com/conda-forge/miniforge#mambaforge
[scverse discourse]: https://discourse.scverse.org/
[issue tracker]: https://github.com/theislab/mapqc/issues
[tests]: https://github.com/theislab/mapqc/actions/workflows/test.yml
[documentation]: https://mapqc.readthedocs.io
[changelog]: https://mapqc.readthedocs.io/en/latest/changelog.html
[api documentation]: https://mapqc.readthedocs.io/en/latest/api.html
[pypi]: https://pypi.org/project/mapqc