https://github.com/tirthajyoti/mlr
Multiple linear regression with statistical inference, residual analysis, direct CSV loading, and other features
https://github.com/tirthajyoti/mlr
analytics data-analytics data-science linear-regression machine-learning modeling predictive-modeling python regression scikit-learn statiscal-learning statistical-analysis statistics
Last synced: 3 months ago
JSON representation
Multiple linear regression with statistical inference, residual analysis, direct CSV loading, and other features
- Host: GitHub
- URL: https://github.com/tirthajyoti/mlr
- Owner: tirthajyoti
- License: gpl-3.0
- Created: 2019-07-31T23:55:41.000Z (almost 6 years ago)
- Default Branch: master
- Last Pushed: 2019-08-12T03:11:02.000Z (almost 6 years ago)
- Last Synced: 2025-03-24T10:12:21.314Z (3 months ago)
- Topics: analytics, data-analytics, data-science, linear-regression, machine-learning, modeling, predictive-modeling, python, regression, scikit-learn, statiscal-learning, statistical-analysis, statistics
- Language: Python
- Homepage: https://mlr.readthedocs.io/en/latest/
- Size: 563 KB
- Stars: 33
- Watchers: 2
- Forks: 10
- Open Issues: 0
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
README
# mlr (`pip install mlr`)
A lightweight, easy-to-use Python package that combines the `scikit-learn`-like simple API with the power of **statistical inference tests**, **visual residual analysis**, **outlier visualization**, **multicollinearity test**, found in packages like `statsmodels` and R language.
Authored and maintained by **Dr. Tirthajyoti Sarkar ([Website](https://tirthajyoti.github.io), [LinkedIn profile](https://www.linkedin.com/in/tirthajyoti-sarkar-2127aa7/))**
### Useful regression metrics,
* MSE, SSE, SST
* R^2, Adjusted R^2
* AIC (Akaike Information Criterion), and BIC (Bayesian Information Criterion)
### Inferential statistics,
* Standard errors
* Confidence intervals
* p-values
* t-test values
* F-statistic
### Visual residual analysis,
* Plots of fitted vs. features,
* Plot of fitted vs. residuals,
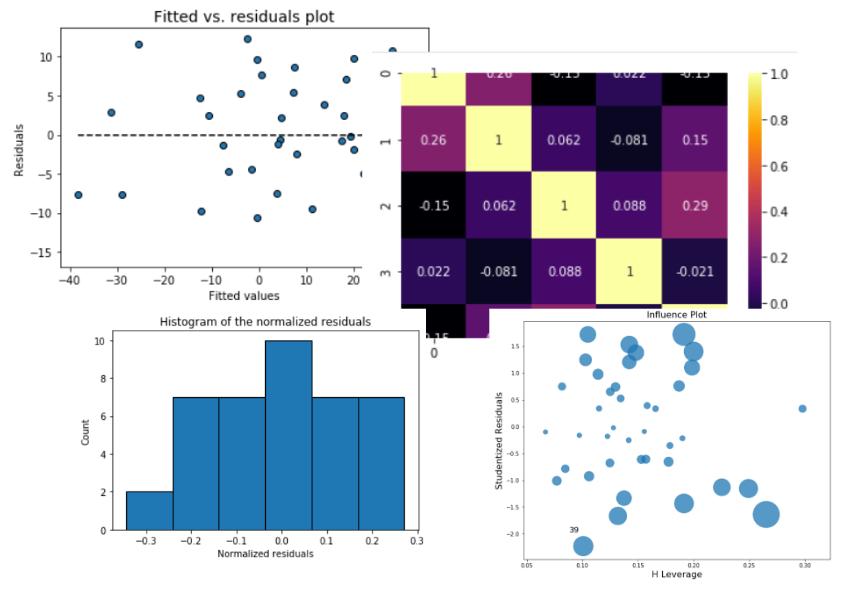
* Histogram of standardized residuals
* Q-Q plot of standardized residuals
### Outlier detection
* Influence plot
* Cook's distance plot
### Multicollinearity
* Pairplot
* Variance infletion factors (VIF)
* Covariance matrix
* Correlation matrix
* Correlation matrix heatmap
## Requirements
* numpy (`pip install numpy`)
* pandas (`pip install pandas`)
* matplotlib (`pip install matplotlib`)
* seaborn (`pip install seaborn`)
* scipy (`pip install scipy`)
* statsmodels (`pip install statsmodels`)
## Install
(On Linux and Windows) You can use ``pip``
```pip install mlr```
(On Mac OS), first install pip,
```
curl https://bootstrap.pypa.io/get-pip.py -o get-pip.py
python get-pip.py
```
Then proceed as above.
For the latest additions, you can always clone this Github repo and run the setup.py.
---
## Quick Start
Import the `MyLinearRegression` class,
```
from mlr.MLR import MyLinearRegression as mlr
import numpy as np
```
Generate some random data
```
num_samples=40
num_dim = 5
X = 10*np.random.random(size=(num_samples,num_dim))
coeff = np.array([2,-3.5,1.2,4.1,-2.5])
y = np.dot(coeff,X.T)+10*np.random.randn(num_samples)
```
Make a model instance,
```
model = mlr()
```
Ingest the data
```
model.ingest_data(X,y)
```
Fit,
```
model.fit()
```
---
## Directly read from a Pandas DataFrame
You can read directly from a Pandas DataFrame. Just give the features/predictors' column names as a list and the target column name as a string to the `fit_dataframe` method.
At this point, only numerical features/targets are supported but in future releases we will support categorical variables too.
```
<... obtain a Pandas DataFrame by some processing>
df = pd.DataFrame(...)
feature_cols = ['X1','X2','X3']
target_col = 'output'
model = mlr()
model.fit_dataframe(X=feature_cols,y = target_col,dataframe=df)
```
---
## Metrics
So far, it looks similar to the linear regression estimator of Scikit-Learn, doesn't it?
Here comes the difference,
### Print all kinds of regression model metrics, one by one,
```
print ("R-squared: ",model.r_squared())
print ("Adjusted R-squared: ",model.adj_r_squared())
print("MSE: ",model.mse())
>> R-squared: 0.8344327025902752
Adjusted R-squared: 0.8100845706182569
MSE: 72.2107655649954
```
### Or, print all the metrics at once!
```
model.print_metrics()
>> sse: 2888.4306
sst: 17445.6591
mse: 72.2108
r^2: 0.8344
adj_r^2: 0.8101
AIC: 296.6986
BIC: 306.8319
```
---
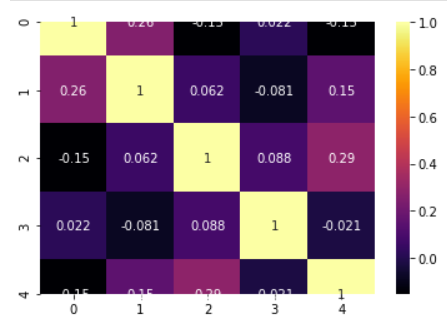
## Correlation matrix, heatmap, covariance
We can build the correlation matrix right after ingesting the data. This matrix gives us an indication how much multicollinearity is present among the features/predictors.
### Correlation matrix
```
model.ingest_data(X,y)
model.corrcoef()
>> array([[ 1. , 0.18424447, -0.00207883, 0.144186 , 0.08678109],
[ 0.18424447, 1. , -0.08098705, -0.05782733, 0.19119872],
[-0.00207883, -0.08098705, 1. , 0.03602977, -0.17560097],
[ 0.144186 , -0.05782733, 0.03602977, 1. , 0.05216212],
[ 0.08678109, 0.19119872, -0.17560097, 0.05216212, 1. ]])
```
### Covariance
```
model.covar()
>> array([[10.28752086, 1.51237819, -0.01770701, 1.47414685, 0.79121778],
[ 1.51237819, 6.54969628, -0.5504233 , -0.47174359, 1.39094876],
[-0.01770701, -0.5504233 , 7.05247111, 0.30499622, -1.32560195],
[ 1.47414685, -0.47174359, 0.30499622, 10.16072256, 0.47264283],
[ 0.79121778, 1.39094876, -1.32560195, 0.47264283, 8.08036806]])
```
### Correlation heatmap
```
model.corrplot(cmap='inferno',annot=True)
```
## Statistical inference
### Perform the F-test of overall significance
It retunrs the F-statistic and the p-value of the test.
If the p-value is a small number you can reject the Null hypothesis that all the regression coefficient is zero. That means a small p-value (generally < 0.01) indicates that the overall regression is statistically significant.
```
model.ftest()
>> (34.270912591948814, 2.3986657277649282e-12)
```
### How about p-values, t-test statistics, and standard errors of the coefficients?
Standard errors and corresponding t-tests give us the p-values for each regression coefficient, which tells us whether that particular coefficient is statistically significant or not (based on the given data).
```
print("P-values:",model.pvalues())
print("t-test values:",model.tvalues())
print("Standard errors:",model.std_err())
>> P-values: [8.33674608e-01 3.27039586e-03 3.80572234e-05 2.59322037e-01 9.95094748e-11 2.82226752e-06]
t-test values: [ 0.21161008 3.1641696 -4.73263963 1.14716519 9.18010412 -5.60342256]
Standard errors: [5.69360847 0.47462621 0.59980706 0.56580141 0.47081187 0.5381103 ]
```
### Confidence intervals
```
model.conf_int()
>> array([[-10.36597959, 12.77562953],
[ 0.53724132, 2.46635435],
[ -4.05762528, -1.61971606],
[ -0.50077913, 1.79891449],
[ 3.36529718, 5.27890687],
[ -4.10883113, -1.92168771]])
```
## Visual analysis of the residuals
Residual analysis is crucial to check the assumptions of a linear regression model. `mlr` helps you check those assumption easily by providing straight-forward visual analytis methods for the residuals.
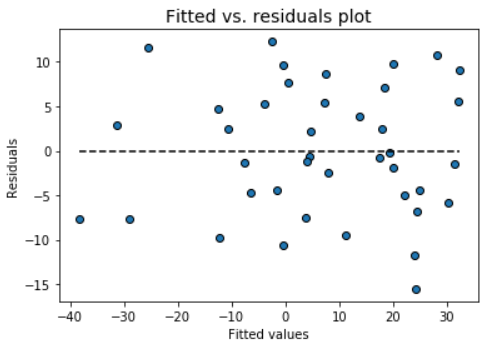
### Fitted vs. residuals plot
Check the assumption of constant variance and uncorrelated features (independence) with this plot
```
model.fitted_vs_residual()
```
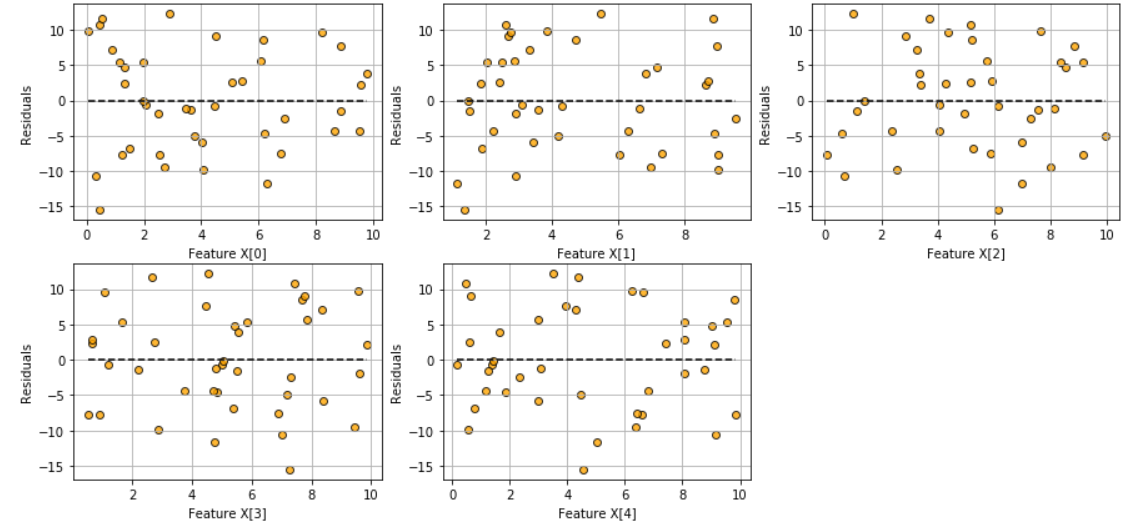
### Fitted vs features plot
Check the assumption of linearity with this plot
```
model.fitted_vs_features()
```
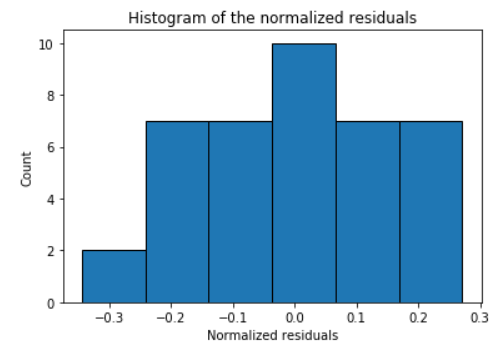
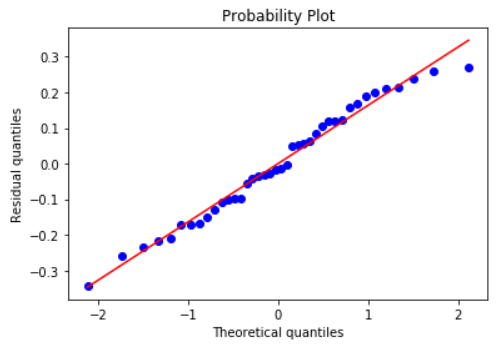
### Histogram and Q-Q plot of standardized residuals
Check the normality assumption of the error terms using these plots,
```
model.histogram_resid()
```
```
model.qqplot_resid()
```
## Do more
Do more fun stuff with your regression model.
More features will be added in the future releases!
* Outlier detection and plots
* Multicollinearity checks