Ecosyste.ms: Awesome
An open API service indexing awesome lists of open source software.
https://github.com/tsudalab/dt-sampler
https://github.com/tsudalab/dt-sampler
Last synced: about 1 month ago
JSON representation
- Host: GitHub
- URL: https://github.com/tsudalab/dt-sampler
- Owner: tsudalab
- Created: 2023-07-17T05:37:51.000Z (over 1 year ago)
- Default Branch: main
- Last Pushed: 2023-09-02T17:42:02.000Z (over 1 year ago)
- Last Synced: 2023-09-03T11:17:41.608Z (over 1 year ago)
- Language: Python
- Size: 144 KB
- Stars: 1
- Watchers: 8
- Forks: 0
- Open Issues: 0
-
Metadata Files:
- Readme: README.md
Awesome Lists containing this project
README
# DT-Sampler
## Details
You can find more details about DT-Sampler at [https://arxiv.org/abs/2307.13333](https://arxiv.org/abs/2307.13333).
## Abstract
DT-sampler is an ensemble model based on decision tree sampling. Different from random forest, DT-sampler uniformly samples decision trees from a given space, which can generate more stable results and provide higher interpretability compared to random forest. DT-sampler only has two key parameters: #node and threshold. #node constrains the size of decision trees generated by DT-sampler and threshold ensures a minimum training accuracy for each decision tree.
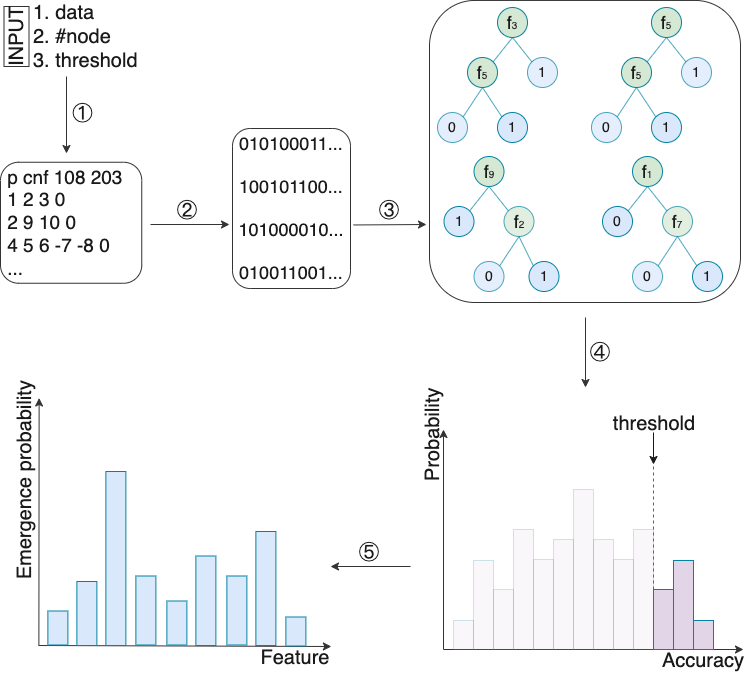
① Encode the construction of decision trees as a SAT problem. \
② Utilize SAT sampler to uniformly sample multiple satisfiable solutions from the high accuracy space.\
③ Decode the satisfiable solutions back into decision trees.\
④ Estimate the training accuracy distribution of the decision trees in the high accuracy space.\
⑤ Measure feature importance by calculating the emergence probability of each feature.
## Requirements
matplotlib == 3.6.3 \
numpy == 1.21.0 \
pandas == 1.5.3 \
pyunigen == 2.5.2 \
scikit_learn == 1.2.1 \
scipy == 1.11.1 \
z3_solver == 4.12.1.0
## Quick Start
```python
...
dt_sampler = DT_sampler(X_train, y_train, #node, threshod, "./cnf/cnf_name.cnf")
dt_sampler.run(#tree, method = "unigen", seed)
...
```
## Contact
Chao Huang ([email protected])\
Department of Computational Biology and Medical Science\
The University of Tokyo