https://github.com/utkuozbulak/adaptive-segmentation-mask-attack
Pre-trained model, code, and materials from the paper "Impact of Adversarial Examples on Deep Learning Models for Biomedical Image Segmentation" (MICCAI 2019).
https://github.com/utkuozbulak/adaptive-segmentation-mask-attack
adversarial-examples segmentation u-net
Last synced: 7 months ago
JSON representation
Pre-trained model, code, and materials from the paper "Impact of Adversarial Examples on Deep Learning Models for Biomedical Image Segmentation" (MICCAI 2019).
- Host: GitHub
- URL: https://github.com/utkuozbulak/adaptive-segmentation-mask-attack
- Owner: utkuozbulak
- License: mit
- Created: 2019-06-28T16:27:34.000Z (over 6 years ago)
- Default Branch: master
- Last Pushed: 2020-07-06T00:18:57.000Z (over 5 years ago)
- Last Synced: 2025-04-19T19:04:53.801Z (8 months ago)
- Topics: adversarial-examples, segmentation, u-net
- Language: Python
- Homepage:
- Size: 10.4 MB
- Stars: 60
- Watchers: 3
- Forks: 10
- Open Issues: 0
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
- Awesome-pytorch-list-CNVersion - Adaptive-segmentation-mask-attack (ASMA)
- Awesome-pytorch-list - Adaptive-segmentation-mask-attack (ASMA)
README
# Adaptive Segmentation Mask Attack
This repository contains the implementation of the _Adaptive Segmentation Mask Attack (ASMA)_, a targeted adversarial example generation method for deep learning segmentation models. This attack was proposed in the paper "_Impact of Adversarial Examples on Deep Learning Models for Biomedical Image Segmentation._" published in the 22nd International Conference on Medical Image Computing and Computer Assisted Intervention, MICCAI-2019. ([Link to the paper](https://arxiv.org/abs/1907.13124))
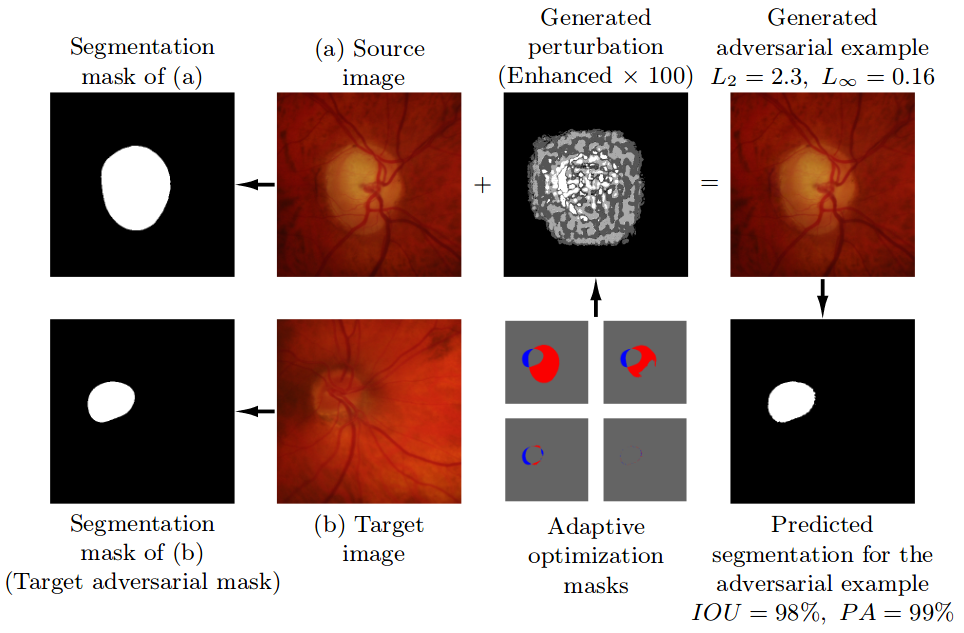
## General Information
This repository is organized as follows:
* **Code** - *src/* folder contains necessary python files to perform the attack and calculate various stats (i.e., correctness and modification)
* **Data** - *data/* folder contains a couple of examples for testing purposes. The data we used in this study can be taken from [1].
* **Model** - Example model used in this repository can be downloaded from https://www.dropbox.com/s/6ziz7s070kkaexp/eye_pretrained_model.pt . _helper_functions.py_ contains a function to load this file and _main.py_ contains an exaple that uses this model.
## Frequently Asked Questions (FAQ)
* How can I run the demo?
**1-** Download the model from https://www.dropbox.com/s/6ziz7s070kkaexp/eye_pretrained_model.pt
**2-** Create a folder called _model_ on the same level as _data_ and _src_, put the model under this (_model_) folder.
**3-** Run _main.py_.
* Would this attack work in multi-class segmentation models?
Yes, given that you provide a proper target mask, model etc.
* Does the code require any modifications in order to make it work for multi-class segmentation models?
No (probably, depending on your model/input). At least the attack itself (adaptive_attack.py) should not require major modifications on its logic.
## Citation
If you find the code in this repository useful for your research, consider citing our paper. Also, feel free to use any visuals available here.
@inproceedings{ozbulak2019impact,
title={Impact of Adversarial Examples on Deep Learning Models for Biomedical Image Segmentation},
author={Ozbulak, Utku and Van Messem, Arnout and De Neve, Wesley},
booktitle={International Conference on Medical Image Computing and Computer-Assisted Intervention},
pages={300--308},
year={2019},
organization={Springer}
}
## Requirements
```
python > 3.5
torch >= 0.4.0
torchvision >= 0.1.9
numpy >= 1.13.0
PIL >= 1.1.7
```
## References
[1] Pena-Betancor C., Gonzalez-Hernandez M., Fumero-Batista F., Sigut J., Medina-Mesa E., Alayon S., Gonzalez M. _Estimation of the relative amount of hemoglobin in the cup and neuroretinal rim using stereoscopic color fundus images._
[2] Ronneberger, O., Fischer, P., Brox, T. _U-Net: Convolutional networks for biomedical image segmentation._