https://github.com/vjcitn/cmg_eye
CMG_EYE repo for AnVIL days demonstrations
https://github.com/vjcitn/cmg_eye
Last synced: 3 months ago
JSON representation
CMG_EYE repo for AnVIL days demonstrations
- Host: GitHub
- URL: https://github.com/vjcitn/cmg_eye
- Owner: vjcitn
- Created: 2021-01-20T21:40:04.000Z (over 4 years ago)
- Default Branch: main
- Last Pushed: 2021-01-26T19:59:51.000Z (over 4 years ago)
- Last Synced: 2025-01-09T13:46:37.657Z (5 months ago)
- Language: R
- Size: 497 KB
- Stars: 0
- Watchers: 3
- Forks: 0
- Open Issues: 0
-
Metadata Files:
- Readme: README.md
Awesome Lists containing this project
README
# CMG_EYE
CMG_EYE repo for AnVIL days demonstrations
Here's the 'dashboard description' for the AnVIL workspace
### Introduction
This workspace was produced to illustrate exploration of CMG data in AnVIL using
- **RStudio** to control all interactions
- **version control with git** for all textual material related to the computations and the workspace
- an **Rmarkdown document** specifying all aspects of
- software extensions required
- data acquisition using the **AnVIL package** for Bioconductor
- interactive tabulation and visualization
- a **shiny app** that explores available FHIR documents on a subset of CMG sequencing data
### Resources and activities
- https://github.com/vjcitn/CMG_EYE is a github repository with `cmgeye.Rmd` and `fhirinspect.R`. The following steps
will help you import the content from this github repository into your own AnVIL session/workspace.
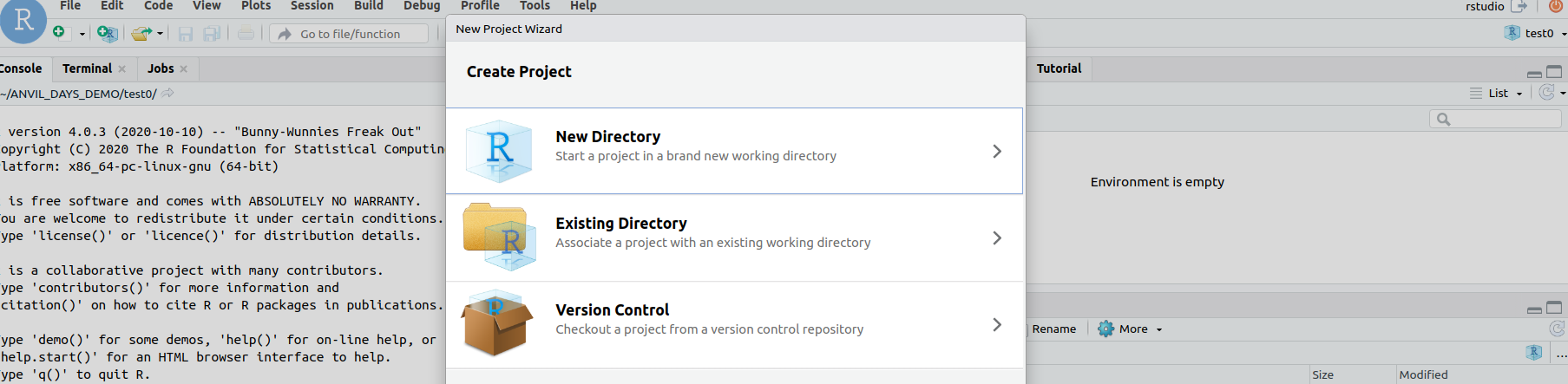
- use Rstudio's `File/New project/` widget
to select a version controlled project
with git
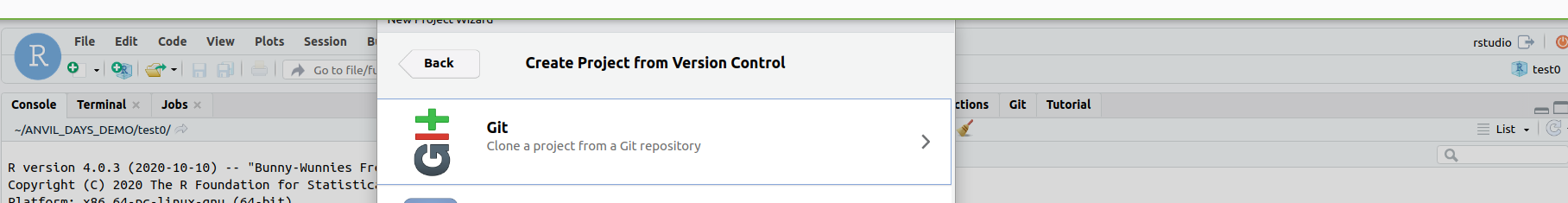
Fill in the git widget
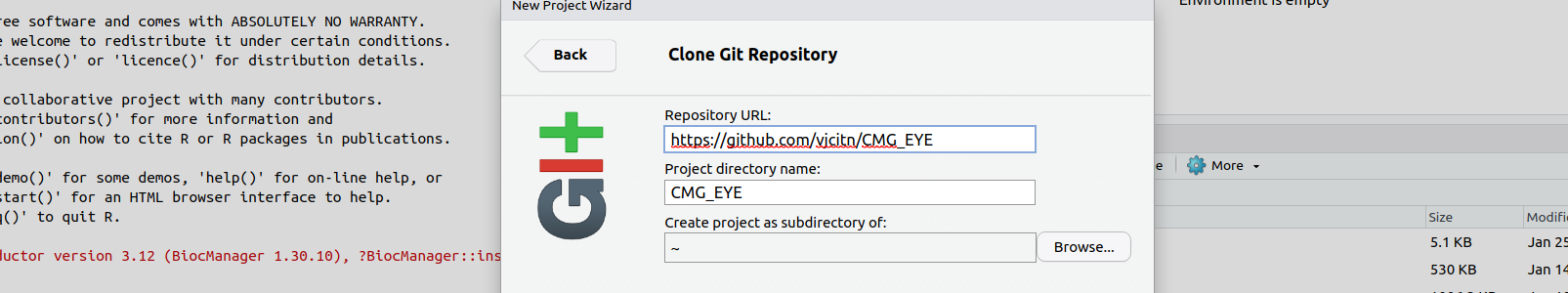
AnVIL RStudio will run git commands to populate a folder of your workspace's disk
### Compile `cmgeye.Rmd`
To begin, use the Rstudio console to run `BiocManager::install("BiocStyle")` as it
is useful for formatting the document we are aiming for.
After installation of BiocStyle completes, click on the `cmgeye.Rmd` file in the Files pane, and then in the upper left pane,
use the `Knit to html_document` widget to generate and view HTML reporting
on the CMG data.
Some commands may require AnVIL_DEV group membership. Set the chunks
involving DRS resolution to eval=FALSE if an error related to permissions crops up.
### Use the `fhirinspector` app
These commands may not work if you do not have
access to `gs://bioc-anvildays-2021`. Email stvjc
at channing.harvard.edu if you are interested and authorized
to work with this information.
Use the commands
```
source("setup_fhirapp.R")
setup_fhirapp(path=tempdir())
```
to copy and unzip a file of FHIR JSON.
These can then be explored with
```
source("fhirinspect.R")
fhirinspector(paste0(tempdir(), "/tmp/CMG"))
```
### The gen3 export process
We've used `gen3.theanvil.io/explorer` to request data on 35 subjects from CMG:
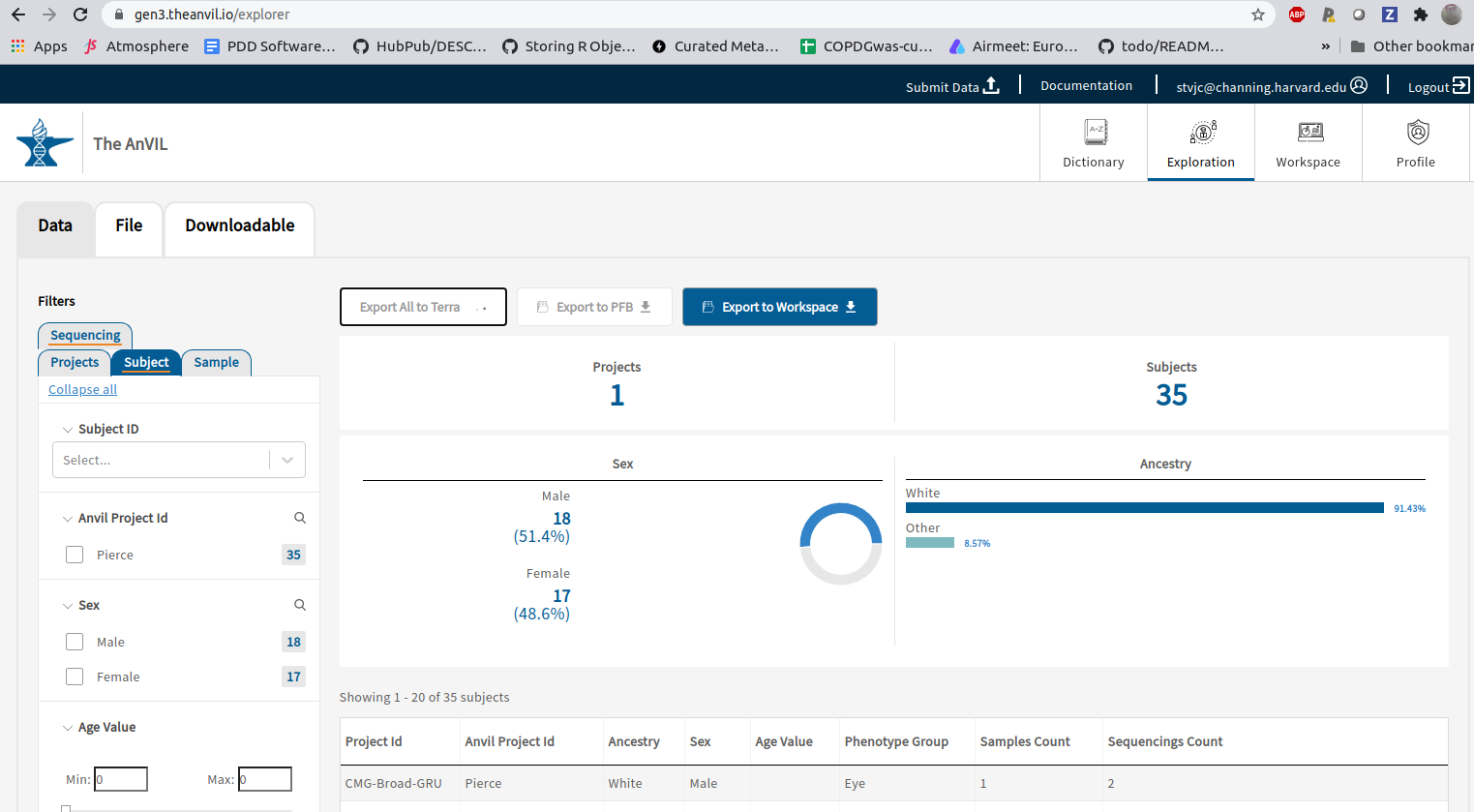
These were exported to AnVIL via the first tab, leading to a "data view" in this workspace (selecting 'subjects'):
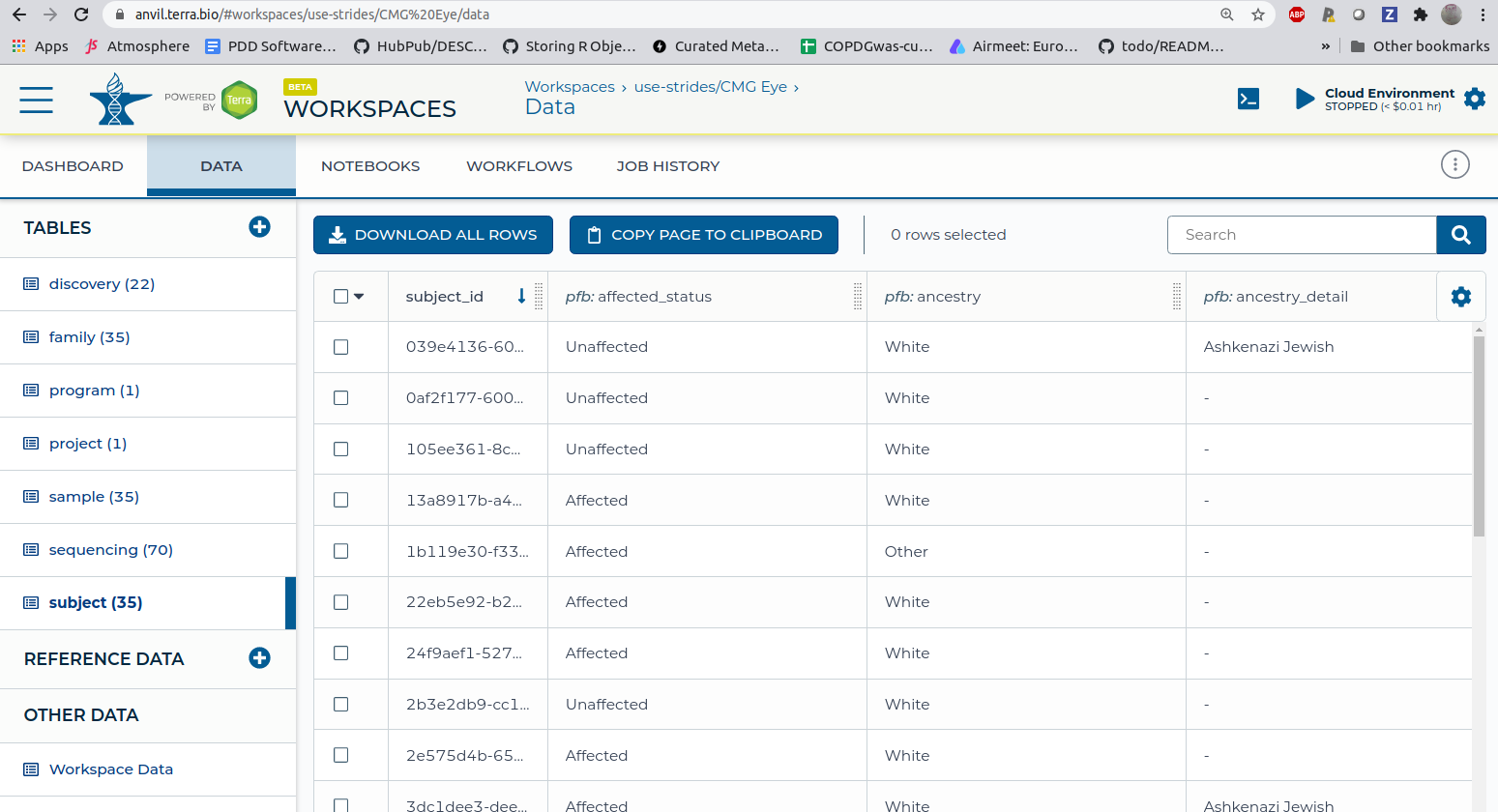
This data view includes DRS identifiers for CRAM files resulting from WGS. The phenotypes are variously
annotated, in multiple fields, with terms 'retinitis pigmentosa', 'RETINITIS PIGMENTOSA 69; RP69',
'Joubert syndrome 1'. OMIM codes are often provided.
We can also use the Gen3 system to generate PFB (portable format for bioinformatics) using the second
tab in the Gen3 explorer page. This is returned as an Avro file, which can be parsed using sparklyr into
a `spark_tbl` instance.