https://github.com/woctezuma/playing-with-simpleitk
Playing with SimpleITK and nnU-Net to process data from the CHAOS challenge on Google Colab.
https://github.com/woctezuma/playing-with-simpleitk
chaos colab colab-notebook colab-notebooks colaboratory colabs nn-unet nn-unet-v2 nn-unetv2 nnu-net nnu-net-v2 nnu-netv2 nnunet nnunet-v2 nnunetv2 segmentation simpleitk sitk
Last synced: 3 months ago
JSON representation
Playing with SimpleITK and nnU-Net to process data from the CHAOS challenge on Google Colab.
- Host: GitHub
- URL: https://github.com/woctezuma/playing-with-simpleitk
- Owner: woctezuma
- License: mit
- Created: 2019-11-25T17:58:52.000Z (almost 6 years ago)
- Default Branch: master
- Last Pushed: 2024-11-14T20:10:25.000Z (11 months ago)
- Last Synced: 2025-04-08T10:51:37.946Z (6 months ago)
- Topics: chaos, colab, colab-notebook, colab-notebooks, colaboratory, colabs, nn-unet, nn-unet-v2, nn-unetv2, nnu-net, nnu-net-v2, nnu-netv2, nnunet, nnunet-v2, nnunetv2, segmentation, simpleitk, sitk
- Language: Jupyter Notebook
- Homepage:
- Size: 29.6 MB
- Stars: 10
- Watchers: 2
- Forks: 7
- Open Issues: 2
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
README
# Playing with SimpleITK and nnU-Net
[![Build status][build-image]][build]
[![Code coverage][codecov-image]][codecov]
[![Code Quality][codacy-image]][codacy]
This repository contains Python code to play with [SimpleITK](https://github.com/SimpleITK/SimpleITK) and [nnU-Net](https://github.com/MIC-DKFZ/nnUNet).
## Requirements
- Install [Python 3.X](https://www.python.org/downloads/),
- Install the required packages:
```bash
pip install -r requirements.txt
```
## Data
- Download data from the [Combined (CT-MR) Healthy Abdominal Organ Segmentation](https://chaos.grand-challenge.org/Combined_Healthy_Abdominal_Organ_Segmentation/) (CHAOS) challenge,
- Extract the archive to `data/`.
Alternatively, data can be found in [**Releases**](https://github.com/woctezuma/playing-with-simpleitk/releases) on Github.
## Usage
### Prepare data
- Convert data from [DICOM](https://en.wikipedia.org/wiki/DICOM) to [NifTI](https://en.wikipedia.org/wiki/Neuroimaging_Informatics_Technology_Initiative) with SimpleITK:
```bash
python convert_to_nii.py
```
- Follow instructions for the folder structure of data, as [detailed for nnU-Net](https://github.com/MIC-DKFZ/nnUNet/blob/master/documentation/dataset_format.md).
### Train nnU-Net
- Run the [`playing_with_pytorch.ipynb`][playing_with_pytorch] notebook.
[![Open In Colab][colab-badge]][playing_with_pytorch]
Training was performed with:
- a training dataset of 5 images downsampled to 4x4x4mm,
- no cross-validation (in which case the validation dataset is the same as the training dataset to find the best checkpoint and to decrease the learning rate).
Training for 1 epoch can require 2m30s, but up to 10m, depending on the machine which you obtained.
Results obtained after ~ 100 epochs are shown below:
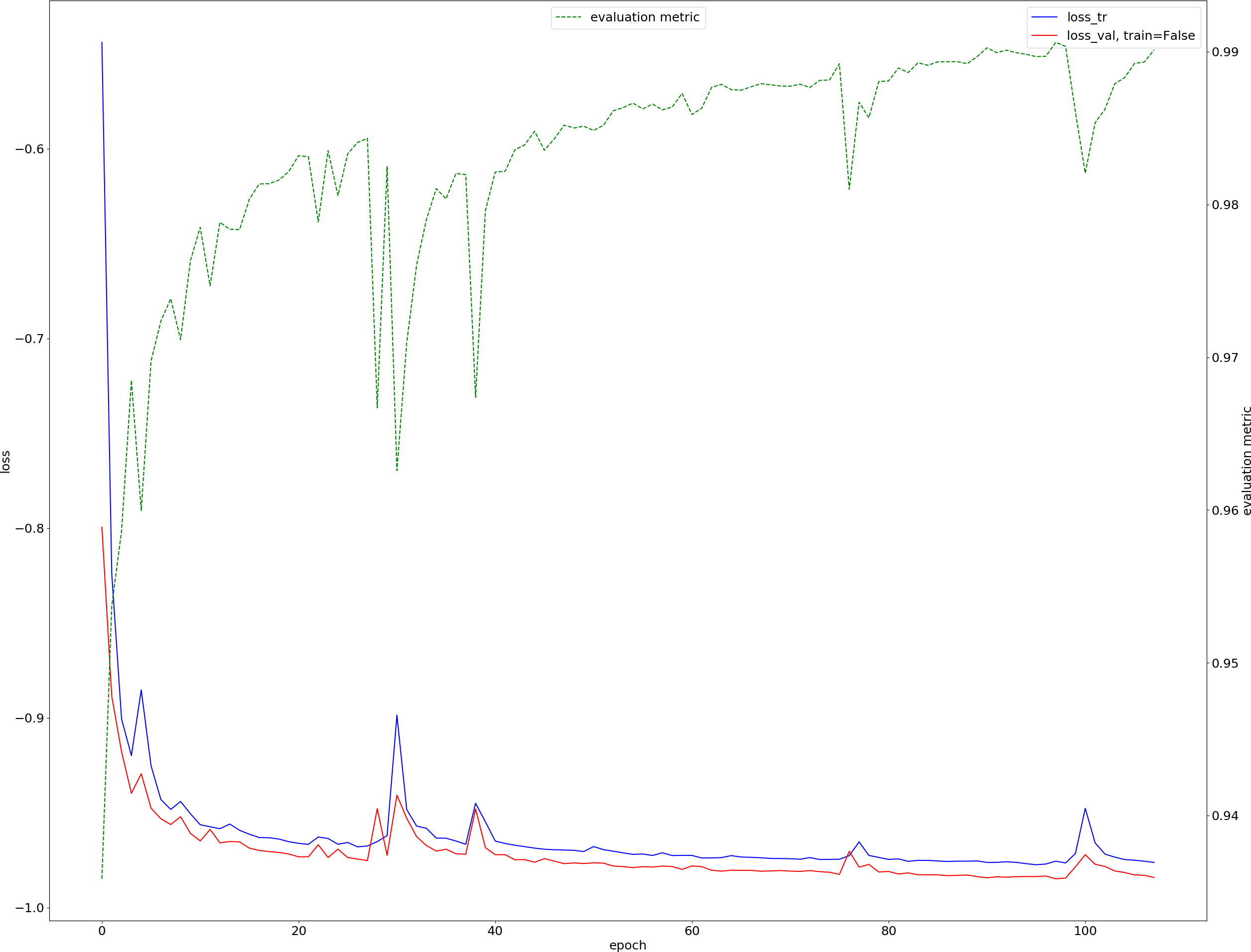
where:
- training loss is in blue,
- validation loss is in red,
- evaluation metric (to maximize) is in green.
In terms of Dice scores, the average is:
- 99.1% for the 5 patients of the training data,
- 94.8% for the 15 patients of the validation data (training data not used).
## References
- [Github repository](https://github.com/SimpleITK/SimpleITK) for SimpleITK,
- [Notebooks](https://github.com/InsightSoftwareConsortium/SimpleITK-Notebooks) for SimpleITK,
- [Documentation](https://simpleitk.readthedocs.io/) for SimpleITK,
- [nnU-Net](https://github.com/MIC-DKFZ/nnUNet),
- [Combined (CT-MR) Healthy Abdominal Organ Segmentation](https://chaos.grand-challenge.org/Combined_Healthy_Abdominal_Organ_Segmentation/) (CHAOS) challenge,
- Evaluation of segmentation results with [tools](https://github.com/emrekavur/CHAOS-evaluation) provided by organizers of CHAOS.
[build]:
[build-image]:
[pyup]:
[dependency-image]:
[python3-image]:
[codecov]:
[codecov-image]:
[codacy]:
[codacy-image]:
[playing_with_pytorch]:
[colab-badge]: