https://github.com/yamaton/vscode-h2o
Add CLI autocomplete functionalities to Shell Script
https://github.com/yamaton/vscode-h2o
bash bioinformatics cli intellisense shellscript vscode-extension
Last synced: 3 months ago
JSON representation
Add CLI autocomplete functionalities to Shell Script
- Host: GitHub
- URL: https://github.com/yamaton/vscode-h2o
- Owner: yamaton
- License: mit
- Created: 2021-05-29T18:01:07.000Z (over 4 years ago)
- Default Branch: main
- Last Pushed: 2023-11-05T23:27:19.000Z (almost 2 years ago)
- Last Synced: 2025-03-28T18:51:58.641Z (7 months ago)
- Topics: bash, bioinformatics, cli, intellisense, shellscript, vscode-extension
- Language: TypeScript
- Homepage:
- Size: 50.2 MB
- Stars: 20
- Watchers: 1
- Forks: 1
- Open Issues: 3
-
Metadata Files:
- Readme: README.md
- Changelog: CHANGELOG.md
- License: LICENSE
Awesome Lists containing this project
README
# Shell Script Command Completion
This extension brings autocompletion and introspection of shell commands to VS Code, enhancing the **Shell Script mode**.
## Features
* Autocomplete command-line options, flags, and subcommands
* Hover to get descriptions for subcommands and options/flags
* **Zero configuration** required
* 🧬 Opt-in support for bioinformatics CLI tools 🧬
## Demos
### Autocomplete in Shell Script
### Introspection with Hover
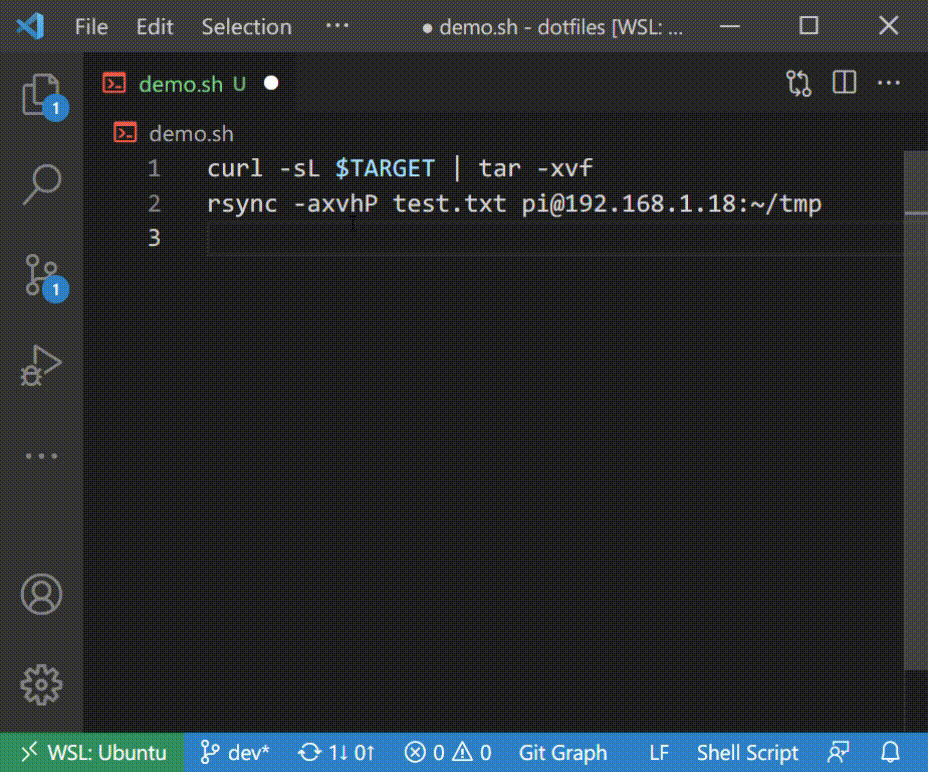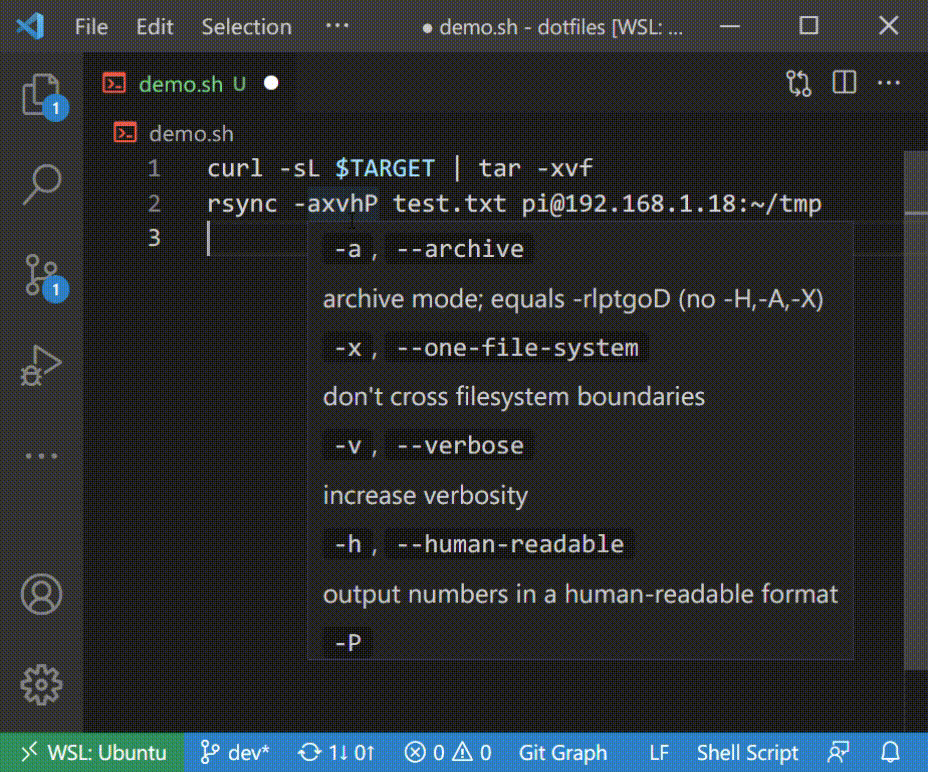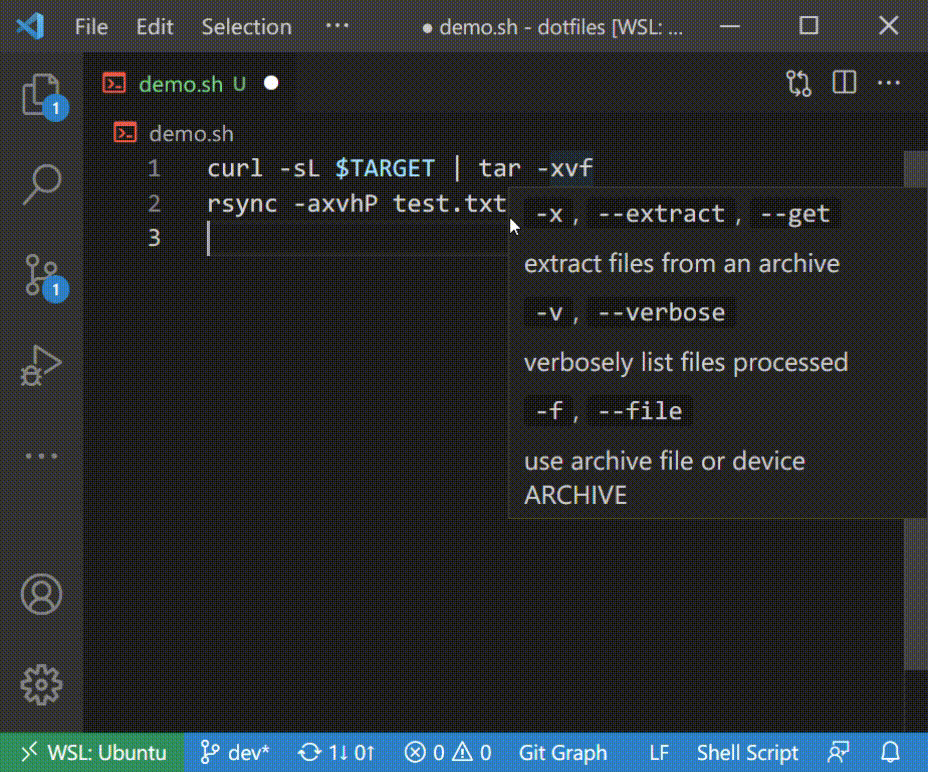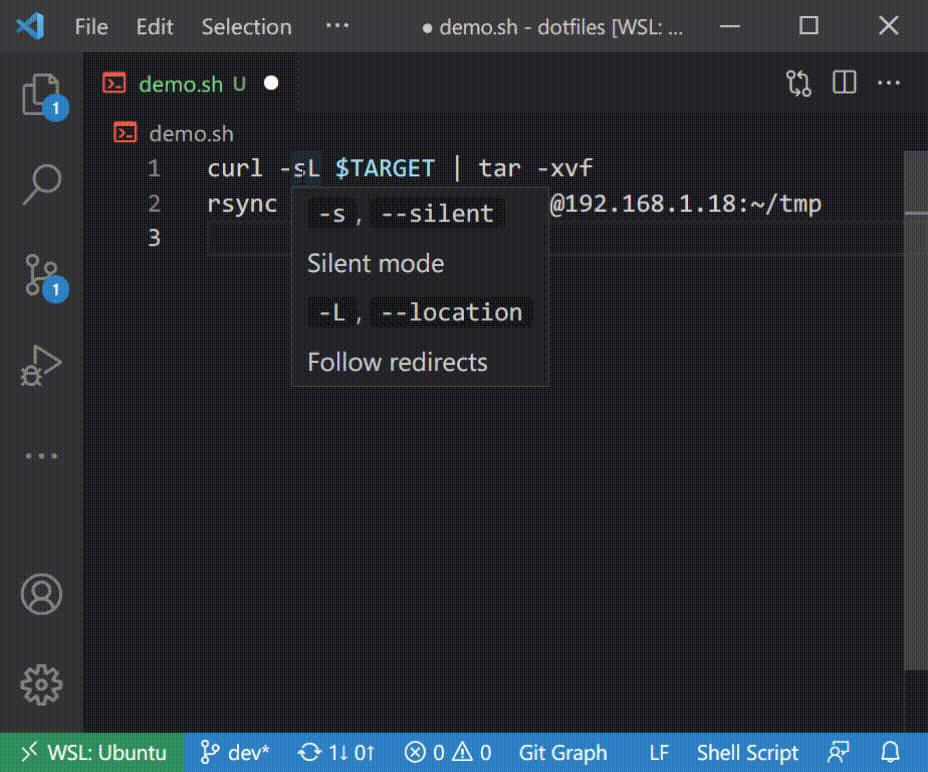
## Supported Commands
The extension comes preloaded with 400+ CLI specifications but can also dynamically create specs by scanning man pages or `--help` documents. The preloaded specs include common tools like `git`, `npm`, `docker`, `terraform`, and many more. See the complete list in [general.txt](https://github.com/yamaton/h2o-curated-data/blob/main/general.txt). If you'd like more tools to be added, please [submit a request here](https://github.com/yamaton/h2o-curated-data/issues/1).
### 🧬 Extra Command Specs for Bioinformatics
Over 500 command specifications for bioinformatics tools can be optionally loaded. In "Shell Script" mode, press `Ctrl`+`Shift`+`P` (or `⌘`+`⇧`+`P` on macOS) and select `Shell Completion: Load Bioinformatics CLI Specs`. If the commands are not recognized, you may need to clear the cache as described below. The supported tools include `BLAST`, `GATK`, `seqkit`, `samtools`, and more. View [bio.txt](https://github.com/yamaton/h2o-curated-data/blob/main/bio.txt) for the full list and [submit any requests for additional tools here](https://github.com/yamaton/h2o-curated-data/issues/1).
## Managing Command Specs
The "Shell Commands" Explorer in the Side Bar displays loaded command specifications.
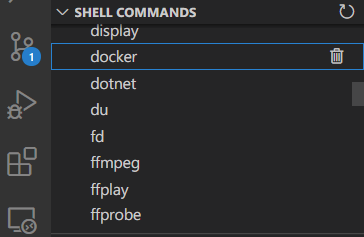
## 🔥 Troubleshooting
### 😞 Not Working?
* If the command is on [this list](https://github.com/yamaton/h2o-curated-data/blob/main/general.txt), activate "Shell Script" mode, then type `Ctrl`+`Shift`+`P` (or `⌘`+`⇧`+`P` on macOS) and choose `Shell Completion: Load Common CLI Specs` to reload the common CLI specs.
* If the command is in [this bio list](https://github.com/yamaton/h2o-curated-data/blob/main/bio.txt), activate "Shell Script" mode, then type `Ctrl`+`Shift`+`P` (or `⌘`+`⇧`+`P` on macOS) and choose `Shell Completion: Load Bioinformatics CLI Specs` to reload the bioinformatics CLI specs.
* If the command is still not recognized, activate "Shell Script" mode, then type `Ctrl`+`Shift`+`P` (or `⌘`+`⇧`+`P` on macOS) and choose `Shell Completion: Remove Command Spec`, then enter the name of the command to remove it from the cache. Our program will then try to recreate the CLI spec.
### 😞 Annoyed by Aggressive Suggestions?
Adjust suggestions with the VS Code settings:
* Suppress **Quick Suggestions**
* Deactivate SPACE-key triggering with **Suggest on Trigger Characters**
Note: These settings apply to other language modes as well.
### 😞 Annoyed by Unwanted Commands?
Use the Shell Commands Explorer to remove unnecessary command specs. To remove all bioinformatics commands, activate "Shell Script" mode, type `Ctrl`+`Shift`+`P` (or `⌘`+`⇧`+`P` on macOS), and choose `Shell Completion: Remove Bioinformatics CLI Specs`.
## 🔧 How the Extension Works
* Utilizes [preprocessed specs](https://github.com/yamaton/h2o-curated-data/tree/main/general/json) when available.
* Extracts CLI information by parsing `man ` or ` --help`.
* Runs on Linux/WSL and macOS only.
* Depends on [tree-sitter](https://tree-sitter.github.io/tree-sitter/) to understand the shell script structure.
## 🛡️ Security with Sandboxing
The extension executes unrecognized commands with `--help` to gather information, potentially posing a risk if untrusted programs are present locally. To mitigate this risk, it uses a sandbox environment if available, ensuring that unrecognized commands run in a controlled and secure environment, limiting network and filesystem access.
* macOS: Always runs in a sandbox with `sandbox-exec`.
* **Linux or WSL**: Consider installing **[bubblewrap](https://wiki.archlinux.org/title/Bubblewrap)**.
## ⚠️ Known Issues
* Autocomplete and hover introspection require either:
* The command in [preprocessed CLI specs](https://github.com/yamaton/h2o-curated-data/tree/main/general/json) to be loaded at startup.
* Successful extraction of CLI information by the included parser from the local environment.