Ecosyste.ms: Awesome
An open API service indexing awesome lists of open source software.
https://github.com/zkamvar/laughing-winner
https://github.com/zkamvar/laughing-winner
Last synced: 2 days ago
JSON representation
- Host: GitHub
- URL: https://github.com/zkamvar/laughing-winner
- Owner: zkamvar
- Created: 2022-07-01T22:34:45.000Z (over 2 years ago)
- Default Branch: main
- Last Pushed: 2022-07-06T00:37:28.000Z (over 2 years ago)
- Last Synced: 2024-06-11T18:23:54.515Z (5 months ago)
- Language: R
- Size: 106 MB
- Stars: 0
- Watchers: 2
- Forks: 1
- Open Issues: 4
-
Metadata Files:
- Readme: README.md
- Contributing: CONTRIBUTING.md
- Funding: .github/FUNDING.yml
- Code of conduct: CODE_OF_CONDUCT.md
Awesome Lists containing this project
README
# Poppr version 2 
[](https://github.com/grunwaldlab/poppr/actions)
## What is *poppr*?
*Poppr* is an R package designed for analysis of populations with mixed modes of
sexual and clonal reproduction. It is built around the framework of [adegenet's](https://github.com/thibautjombart/adegenet/)
genind and genlight objects and offers the following implementations:
- clone censoring of populations at any of multiple levels of a hierarchy
- convenient counting of multilocus genotypes and sub-setting of populations with multiple levels of hierarchy
- define multilocus genotypes
- calculation of indices of genotypic diversity, evenness, richness, and rarefaction
- drawing of dendrograms with bootstrap support for genetic distances
- drawing of minimum spanning networks for genetic distances
- calculation of the index of association
(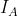 )
)
or (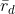 )
)
- batch processing on any server that has R (≥ 2.15.1) installed
- calculation of Bruvo's distance for microsatellite (SSR) markers (implemented in C for speed)
- import of data from and export to [GenAlEx](https://biology-assets.anu.edu.au/GenAlEx/Welcome.html "GenAlEx Homepage")
### New in version 2.0:
- handling of genomic SNP data
- custom multilocus genotype definitions
- collapse multilocus lineages by genetic distance
- calculate reticulate minimum spanning networks
- calculate index of association in a sliding window across snps
- bootstrapping of MLG diversity statistics
- [interactive exploration of minimum spanning networks](https://github.com/zkamvar/poppr_msn_shiny)
- and more!
For full details, see the NEWS file or type in your R console:
```R
news(Version >= "2.0.0", package = "poppr")
```
## Citation
If you use *poppr* at all, please specify the version and cite:
> Kamvar ZN, Tabima JF, Grünwald NJ. (2014) Poppr: an R package for genetic
> analysis of populations with clonal, partially clonal, and/or sexual
> reproduction. PeerJ 2:e281
> [https://doi.org/10.7717/peerj.281](https://doi.org/10.7717/peerj.281)
If you use *poppr* in a presentation please mention it as the *poppr* R package,
specify the version, and use our logo:
[(png)](https://github.com/grunwaldlab/poppr/blob/main/vignettes/jalapeno_logo.png) | [(svg)](https://github.com/grunwaldlab/poppr/blob/main/vignettes/jalapeno_logo.svg).
Additionally, if you use any following functionalities:
- minimum spanning networks with reticulation
- collapsing multilocus genotypes into multilocus lineages with `mlg.filter()`
- custom multilocus genotype definitions with `mlg.custom()`
- index of association for genomic data with `win.ia()` or `samp.ia()`
- bootstrapping any genetic distance with genind, genlight, or genpop objects with `aboot()`
Please also cite:
> Kamvar ZN, Brooks JC and Grünwald NJ (2015) Novel R tools for analysis of
> genome-wide population genetic data with emphasis on clonality. Front. Genet.
> 6:208. doi:
> [10.3389/fgene.2015.00208](https://www.frontiersin.org/articles/10.3389/fgene.2015.00208/abstract)
You can obtain citation information in R by typing:
```R
citation(package = "poppr")
```
## Installation
### From CRAN
[Binary versions for mac and windows are available for R ≥ 2.15.1 **here**](https://cran.r-project.org/package=poppr).
To install, make sure R is at least version 2.15.1 (the authors recommend ≥
3.0), and in your console, type:
```R
install.packages("poppr")
```
If you want the absolute latest version of *poppr*, see about installing
unreleased versions below.
***
### Stable version
New features are occasionally added to {poppr}, but it can take time for it to
get to CRAN. If you know that you want the latest version of {poppr}, (which
will contain bug fixes and new features to be included in future releases), then
you can use the custom R-Universe repository, which is updated hourly:
To install poppr from the R-Universe, you can use the following code:
```R
universe <- c("https://zkamvar.r-universe.dev", "https://cloud.r-project.org")
install.packages("poppr", repos = universe)
```
The universe repository also contain up-to-date versions of {adegenet} and
{hierfstat}, which are commonly used in conjunction with {poppr} and are
notoriously out of date on CRAN.
### Unstable/Development versions
All Development versions of {poppr} will be on GitHub, but need to be compiled.
To install this package from github, make sure you have the following:
- [Xcode](https://developer.apple.com/xcode/) (OSX)
OR [Rtools](https://cran.r-project.org/bin/windows/Rtools/) (Windows)
- [{remotes}](https://github.com/r-lib/remotes) (to install, use: `install.packages("remotes")`)
> For Linux users, make sure that the function `getOption("unzip")` returns
> `"unzip"` or `"internal"`. If it does not, then run `options(unzip =
> "internal")`.
Once you have {remotes} and a C compiler installed, you can use the
`install_github()` function to install the current version from github.
All new features in testing will be released on different branches. These
features will be in various stages of development and may or may not be
documented. Install with caution. The below command would install features on
the branch called "devel". Note that these branches might be out of date
from the main branch. Note: if you don't have LaTeX installed, you should set
`build_vignettes = FALSE`.
```R
remotes::install_github(repo = "grunwaldlab/poppr@devel", build_vignettes = TRUE)
library("poppr")
```
## Help / Documentation
### R documentation
To access a descriptive index of help files in *poppr*, type in your console:
```r
package?poppr
```
### Vignettes
A few vignettes have been written for poppr:
|Title |Command |
|:------------------------------|:-------------------------------------|
|Algorightms and Equations |`vignette("algo", "poppr")` |
|Data import and manipulation |`vignette("poppr_manual", "poppr")` |
|Multilocus Genotype Analysis |`vignette("mlg", "poppr")` |
### User Group
Users who have any questions/comments/suggestions regarding any version of poppr
(stable or development) should direct their comments to the [Poppr google
group](https://groups.google.com/d/forum/poppr)
### Book/Primer
In Spring of 2014, Dr. Niklaus J. Grünwald, Dr. Sydney E. Everhart and Zhian N.
Kamvar wrote a primer for population genetic analysis in R located at
https://grunwaldlab.github.io/Population_Genetics_in_R.
## Contributing
Please note that this project is released with a [Contributor Code of
Conduct](https://github.com/grunwaldlab/poppr/blob/main/CODE_OF_CONDUCT.md). By
participating in this project you agree to abide by its
terms. If you wish to contribute code to *poppr*, please fork the repository and
create a pull request with your added feature.