https://github.com/zsvoboda/dbd
dbd is a database prototyping tool that enables data analysts and engineers to quickly load and transform data in SQL databases.
https://github.com/zsvoboda/dbd
bigquery csv database database-schemas elt etl excel json mysql parquet postgresql python python3 redshift snowflake sql sqlite xls xlsx
Last synced: 5 months ago
JSON representation
dbd is a database prototyping tool that enables data analysts and engineers to quickly load and transform data in SQL databases.
- Host: GitHub
- URL: https://github.com/zsvoboda/dbd
- Owner: zsvoboda
- License: bsd-3-clause
- Created: 2022-01-02T17:28:59.000Z (about 4 years ago)
- Default Branch: master
- Last Pushed: 2022-02-13T18:30:58.000Z (about 4 years ago)
- Last Synced: 2025-09-08T13:58:48.233Z (5 months ago)
- Topics: bigquery, csv, database, database-schemas, elt, etl, excel, json, mysql, parquet, postgresql, python, python3, redshift, snowflake, sql, sqlite, xls, xlsx
- Language: Python
- Homepage:
- Size: 2.81 MB
- Stars: 57
- Watchers: 2
- Forks: 2
- Open Issues: 0
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
- jimsghstars - zsvoboda/dbd - dbd is a database prototyping tool that enables data analysts and engineers to quickly load and transform data in SQL databases. (Python)
README
# dbd: database prototyping tool
dbd is a database prototyping tool that enables data analysts and engineers to quickly load and transform data in SQL databases.
dbd helps you with following tasks:
- Loading CSV, JSON, Excel, and Parquet data to database. It supports both local and online files (HTTP URLs). Data can be loaded incrementally or in full.
- Transforming data in existing database tables using insert-from-sql statements.
- Executing DDL (Data Definition Language) SQL scripts (statements like `CREATE SCHEMA`, etc.).
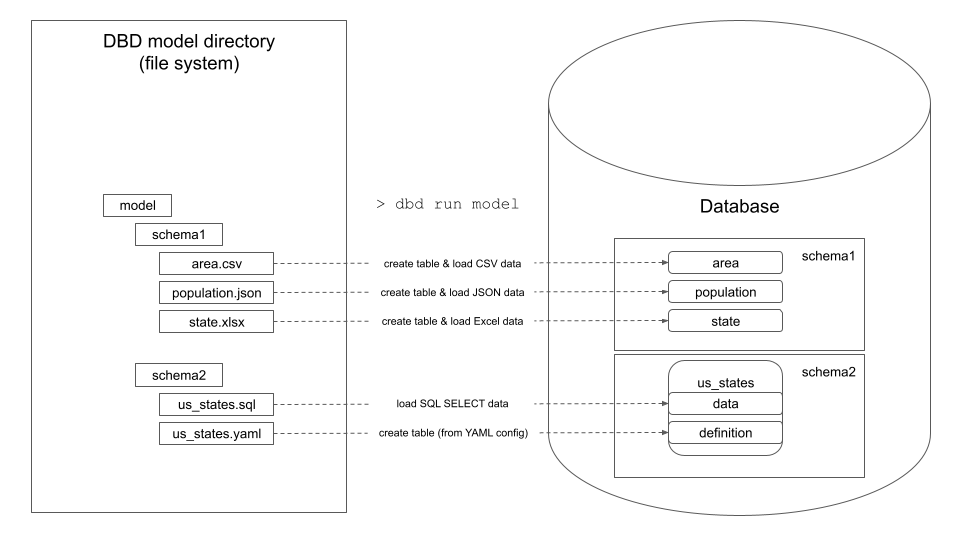
## How dbd works
dbd processes a model directory that contains directories and files:
- **Directories** create new database schemas.
- **Files** create new database table or view. The new table's or view's name is the same as the data file name.
- `.csv`, `.json`, `.xlsx`, and `.parquet` data files are introspected and loaded to database as tables.
- `.sql` files that contain SQL SELECT statements are executed and the result is loaded to database as table or view.
- `.ref` files contain one or more local paths or URLs pointing to supported data files. The referenced files are loaded to database as tables.
- `.yaml` files contain metadata for the files above. The `.yaml` file has the same name as a data, `.sql`, or `.ref` file and specifies details of target table's columns (data types, constraints, indexes, etc.). `.yaml` files are optional. If not specified, dbd uses defaults (e.g. `TEXT` data types for CSV columns)
- `.ddl` files contain multiple SQL statements separated by semicolon that are executed against the database.
dbd knows the correct order in which to process files in the model directory to respect mutual dependencies between
created objects.
dbd currently supports Postgres, MySQL/MariaDB, SQLite, Snowflake, BigQuery, and Redshift databases.
## Getting started and Examples
A short 5-minute getting started tutorial is available
[here](https://zsvoboda.medium.com/analyze-covid-data-in-less-than-5-minutes-9176f440dd1a).
You can also check out dbd's [examples here](https://github.com/zsvoboda/dbd/tree/master/examples).
The easiest way how to execute them is to either clone or download dbd's github repository and start with the
[SQLite examples](https://github.com/zsvoboda/dbd/tree/master/examples/sqlite).
```shell
python3 -m venv dbd-env
source dbd-env/bin/activate
pip3 install dbd
git clone https://github.com/zsvoboda/dbd.git
cd dbd/examples/sqlite/basic
dbd run .
```
These commands should create a new `basic.db` SQLite database with `area`, `population`, and `state` tables that are created and loaded from the corresponding files in the `model` directory.
## Installing dbd
dbd requires Python 3.8 or higher.
### Prerequisites
Check that you have a recent version of Python 3.8 or higher.
```shell
python3 -V
```
if not use a package manager to install the latest python:
On Fedora run:
```shell
sudo yum install python3
```
On Ubuntu run:
```shell
sudo apt install python3
```
Install Python virtual environment:
On Fedora run:
```shell
sudo yum install python3-virtualenv
```
On Ubuntu run:
```shell
sudo apt install python3-venv
```
On Windows just install Python 3.8 or higher from the Store.
Then activate the virtual environment:
On Linux run:
```shell
python3 -m venv dbd-env
source dbd-env/bin/activate
```
On Windows run:
```shell
python3 -m venv dbd-env
call dbd-env\Scripts\activate.bat
```
### PyPI
```shell
pip3 install dbd
```
OR
```shell
git clone https://github.com/zsvoboda/dbd.git
cd dbd
pip3 install .
```
### Running dbd
`dbd` installs a command line executable that must reside on your path. Sometimes Python places the executable
(called `dbd`) outside of your `PATH`. Try to execute `dbd` after the installation. If the command cannot be found,
try to execute
```shell
export PATH=~/.local/bin:$PATH
```
and run `dbd` again. `pip3` usually complains about the fact that the directory where it is placing the executable is
not in `PATH`. You need to take the scripts directory that it suggests and put it on your `PATH`.
Once you can execute the `dbd` command, clone the dbd repository and start with the SQLite examples:
```shell
git clone https://github.com/zsvoboda/dbd.git
cd dbd/examples/sqlite/basic
dbd run .
```
You can also start with [this tutorial](https://zsvoboda.medium.com/analyze-covid-data-in-less-than-5-minutes-9176f440dd1a).
## Starting a new dbd project
You can generate dbd project initial layout by executing `init` command:
```shell
dbd init
```
The `init` command generates a new dbd project directory with the following content:
- `model` directory that contains the content files.
- `dbd.profile` configuration file that defines database connections. The profile file is usually shared by more dbd projects.
- `dbd.project` project configuration file references one of the connections from the profile file and define the `model` directory location.
## dbd profile configuration file
dbd stores database connections in the `dbd.profile` configuration file. dbd searches for it in the current directory or in your home directory. You can use `--profile` option to point it to a profile file in different location.
The profile file is YAML file with the following structure:
```yaml
databases:
db1:
db.url:
db2:
db.url:
db3:
db.url:
```
Read [this document](https://docs.sqlalchemy.org/en/14/core/engines.html) for more details about specific SQLAlchemy database URL formats.
## dbd project configuration file
dbd stores project configuration in project configuration file that is usually stored in your dbd project directory. dbd searches for `dbd.project` file in your project's directory root. You can also use the `--project` option of the `dbd` command to specify a custom project configuration file.
The project configuration file also uses YAML format and references dbd model directory and databse connection from a profile config file. All paths in project file are either absolute or relative to the directory where the profile file is located.
For example:
```yaml
model: ./model
database: db2
```
## Model directory
Model directory contains directories and files. Directories represent database schemas. Files, in most cases, represent database tables.
For example, this `model` directory layout
```text
dbd-model-directory
+- schema1
+-- us_states.csv
+- schema2
+-- us_counties.csv
```
creates two database schemas: `schema1` and `schema2` and two database tables: `us_states` in `schema1` and `us_counties` in `schema2`. Both tables are populated with the data from the CSV files.
dbd supports following files located in the `model` directory:
* __DATA files:__ `.csv`, `.json`, `.xls`, `.xlsx`, `.parquet` files are loaded to database as tables
* __REF files:__ `.ref` files contain one or more absolute or relative paths to local files or URLs of online data files that are loaded to database as tables. All referenced files must have the same structure as they are loaded to the same table.
* __SQL files:__ `.sql` with SQL SELECT statements are executed using insert-from-select SQL construct. The INSERT command is generated (the SQL file only contains a SQL SELECT statement)
* __DDL files:__ contain a sequence of SQL statements separated by semicolon. The DDL files can be named `prolog.ddl` and `epilog.ddl`. The `prolog.ddl` is executed before all other files in a specific schema. The `epilog.ddl` is executed last. The `prolog.ddl` and `epilog.ddl` in the top-level model directory are executed as the very first or the very last files in the model.
* __YAML files:__ specify additional configuration for the __DATA__, __SQL__, and __REF__ files.
## REF files
`.ref` file contains one or more references to files that dbd loads to the database as tables. The references can be URLs, absolute file paths or paths relative to the `.ref` file. All referenced data files must have the same structure as they are loaded to the same database table.
Here is an example of a `.ref` file:
```
https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_daily_reports/01-03-2022.csv
https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_daily_reports/01-04-2022.csv
../data/01-05-2022.csv
../data/01-06-2022.csv
```
The paths and URLs can point to data files with different formats (e.g. CSV or JSON) as long as the files have the
same structure (number of columns and column types).
### Referencing files inside ZIP archives
REF files support paths that reference files inside ZIP archives using the `>` path separator. For example:
`../data/archive.zip>covid-variants.csv`
OR
`https://raw.githubusercontent.com/zsvoboda/dbd/master/tests/fixtures/capabilities/zip_local/data/archive.zip>covid-variants.csv`
### Kaggle datasets
You can reference a kaggle dataset using the `kaggle://kaggle-dataset-name>dataset-file` url.
For example:
`kaggle://kalilurrahman/new-york-times-covid19-dataset>us.csv`
#### Kaggle authentication
To use the Kaggle API, sign up for a Kaggle account at https://www.kaggle.com. Then go to the 'Account' tab
of your user profile (`https://www.kaggle.com//account`) and select 'Create API Token'.
This will trigger the download of kaggle.json, a file containing your API credentials.
Place this file in the location `~/.kaggle/kaggle.json` (on Windows in the location
`C:\Users\\.kaggle\kaggle.json` - you can check the exact location,
sans drive, with `echo %HOMEPATH%`).
You can define a shell environment variable `KAGGLE_CONFIG_DIR` to change this location to `$KAGGLE_CONFIG_DIR/kaggle.json`
(on Windows it will be `%KAGGLE_CONFIG_DIR%\kaggle.json`).
For your security, ensure that other users of your computer do not have read access to your credentials.
On Unix-based systems you can do this with the following command:
`chmod 600 ~/.kaggle/kaggle.json`
You can also choose to export your Kaggle username and token to the environment:
```shell
export KAGGLE_USERNAME=datadinosaur
export KAGGLE_KEY=xxxxxxxxxxxxxx
```
## SQL files
`.sql` file performs SQL data transformation in the target database. It contains a SQL SELECT statement that
dbd wraps in insert-from-select statement, executes it, and stores the result into a table or view that inherits its name from the SQL file name.
Here is an example of `us_states.sql` file that creates a new `us_states` database table:
```sqlite
SELECT
state.abbrev AS state_code,
state.state AS state_name,
population.population AS state_population,
area.area_sq_mi AS state_area_sq_mi
FROM state
JOIN population ON population.state = state.abbrev
JOIN area on area.state_name = state.state
```
## YAML files
`.yaml` file specifies additional configuration for a corresponding __DATA__, __REF__ or __SQL__ file with the same base file name. Here is a YAML configuration example for the `us_states.sql` file above:
```yaml
table:
columns:
state_code:
nullable: false
primary_key: true
type: CHAR(2)
state_name:
nullable: false
index: true
type: VARCHAR(50)
state_population:
nullable: false
type: INTEGER
state_area_sq_mi:
nullable: false
type: INTEGER
process:
materialization: table
mode: drop
```
This `.yaml` file re-types the `state_population` and the `state_area_sq_mi` columns to INTEGER, disallows NULL values in all columns, and makes the `state_code` column table's primary key.
You don't have to describe all table's columns. The columns that you leave out will have their types
set to the default TEXT datatype in case of DATA files and is defined by the insert-from-select in case of SQL files.
The `us_states.sql` table is dropped and data are re-loaded in full everytime the dbd executes this model.
### Table section
`.yaml` file's columns are mapped to a columns of the table that dbd creates from a corresponding __DATA__, __REF__ or __SQL__ file. For example, a CSV header columns or SQL SELECT column `AS` column clauses.
dbd supports following column's parameters:
* __type:__ column's SQL type.
* __primary_key:__ is the column part of table's primary key (true|false)?
* __foreign_keys:__ all other database table columns that are referenced from a column in table (in format `foreign-table`.`referenced-column`).
* __nullable:__ does column allow null values (true|false)?
* __index:__ is column indexed (true|false)?
* __unique:__ does column store unique values (true|false)?
### Process section
The `process` section defines following processing options:
* __materialization:__ specifies whether dbd creates a physical `table` or a `view` when processing SQL file. The __REF__ and __DATA__ files always yield physical table.
* __mode:__ specifies what dbd does with table's data. You can specify values `drop`, `truncate`, or `keep`. The __mode__ option is ignored for views.
## Iterative development
The `dbd` tool's parameter `--only` helps with iterative development process by allowing you to specify a subset of the
tables to process. The `--only` parameter accepts a comma-separated list of fully qualified table names (`schema`.`table-name`).
For example:
`dbd run --only stage.ext_country`
only processes the `ext_country` table in the `stage` schema.
The `--only` parameter also processes all passed table's dependencies. You can skip the dependencies with `--no-deps` option.
## Jinja templates
Most of model files support [Jinja2 templates](https://jinja.palletsprojects.com/en/3.0.x/). For example, this __REF__ file loads 6 CSV files to database (4 online files from a URL and 2 from a local filesystem):
```jinja
{% for n in range(4) %}
https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_daily_reports/01-0{{ n+1 }}-2022.csv
{% endfor %}
../data/01-05-2022.csv
../data/01-06-2022.csv
```
Profile an project configuration files also us Jinja2 templates. You can expend any environment variable with the `{{ environment-variable-name }}` syntax.
For example, you can define your database connection parameters like username or password in environment variables and use this profile configuration file:
```yaml
databases:
states_snowflake:
db.url: "snowflake://{{ SNOWFLAKE_USER }}:{{ SNOWFLAKE_PASSWORD }}@{{ SNOWFLAKE_ACCOUNT_IDENTIFIER }}/{{ SNOWFLAKE_DB }}/{{ SNOWFLAKE_SCHEMA }}?warehouse={{SNOWFLAKE_WAREHOUSE }}"
covid:
db.url: "snowflake://{{ SNOWFLAKE_USER }}:{{ SNOWFLAKE_PASSWORD }}@{{ SNOWFLAKE_ACCOUNT_IDENTIFIER }}/{{ SNOWFLAKE_DB }}/{{ SNOWFLAKE_SCHEMA }}?warehouse={{SNOWFLAKE_WAREHOUSE }}"
```
## Fast data loading mode
All supported database engines except SQLite support fast data loading mode. In this mode, data are loaded to a
database table using bulk load (SQL COPY) command instead of individual INSERT statements.
MySQL and Redshift require additional configuration to enable fast data loading mode.
Without this extra configuration dbd reverts to slow inserting mode via INSERT statements.
### MySQL
To enable fast loading mode, you need specify `local_infile=1` query parameter in the MySQL connection url.
You also must enable the LOCAL INFILE mode on your MySQL server. You can for example do this by executing this
SQL statement:
```mysql
SET GLOBAL local_infile = true
```
### Redshift
To enable fast loading mode, you need specify `copy_stage` parameter in the `dbd.project` configuration file.
The `copy_stage` parameter must reference a storage definition in your `dbd.profile` configuration file.
Check the example configuration files in the `examples/redshift/covid_cz` directory. Here are the example definitions of the
environment variables that these configuration files use:
```shell
export AWS_COVID_STAGE_S3_URL="s3://covid/stage"
export AWS_COVID_STAGE_S3_ACCESS_KEY="AKIA43SWERQGXMUYFIGMA"
export AWS_COVID_STAGE_S3_S3_SECRET_KEY="iujI78eDuFFGJF6PSjY/4CIhEJdMNkuS3g4t0BRwX"
```
## License
dbd code is open-sourced under [BSD 3-clause license](LICENSE).
## Resources and References
- [dbd getting started](https://zsvoboda.medium.com/analyze-covid-data-in-less-than-5-minutes-9176f440dd1a)
- [dbd github repo](https://github.com/zsvoboda/dbd)
- [dbd PyPi](https://pypi.org/project/dbd/)
- [Submit issue](https://github.com/zsvoboda/dbd/issues)