https://sydneybiox.github.io/CiteFuse/
CiteFuse:
https://sydneybiox.github.io/CiteFuse/
bioinformatics r single-cell
Last synced: about 2 months ago
JSON representation
CiteFuse:
- Host: GitHub
- URL: https://sydneybiox.github.io/CiteFuse/
- Owner: SydneyBioX
- Created: 2019-11-25T07:37:15.000Z (over 5 years ago)
- Default Branch: master
- Last Pushed: 2023-02-07T04:36:26.000Z (over 2 years ago)
- Last Synced: 2024-11-22T01:15:03.308Z (6 months ago)
- Topics: bioinformatics, r, single-cell
- Language: R
- Homepage: https://sydneybiox.github.io/CiteFuse/index.html
- Size: 105 MB
- Stars: 26
- Watchers: 11
- Forks: 5
- Open Issues: 3
-
Metadata Files:
- Readme: README.md
Awesome Lists containing this project
- awesome-multi-omics - CiteFuse - Kim - CITE-seq data analysis [paper](https://doi.org/10.1093/bioinformatics/btaa282) (Software packages and methods / Single cell multi-omics)
README
# CiteFuse
CiteFuse is a streamlined package consisting of a suite of tools for pre-processing, modality integration, clustering, differential RNA and ADT expression analysis, ADT evaluation, ligand-receptor interaction analysis, and interactive web-based visualization of CITE-seq data
## Installation
Install CiteFuse via Bioconductor
```r
library(BiocManager)
BiocManager::install("CiteFuse")
```
Install github version CiteFuse
```r
BiocManager::install("SydneyBioX/CiteFuse")
```
## Vignette
You can find the vignette at our website: https://sydneybiox.github.io/CiteFuse/articles/CiteFuse.html.
## CiteFuse overview
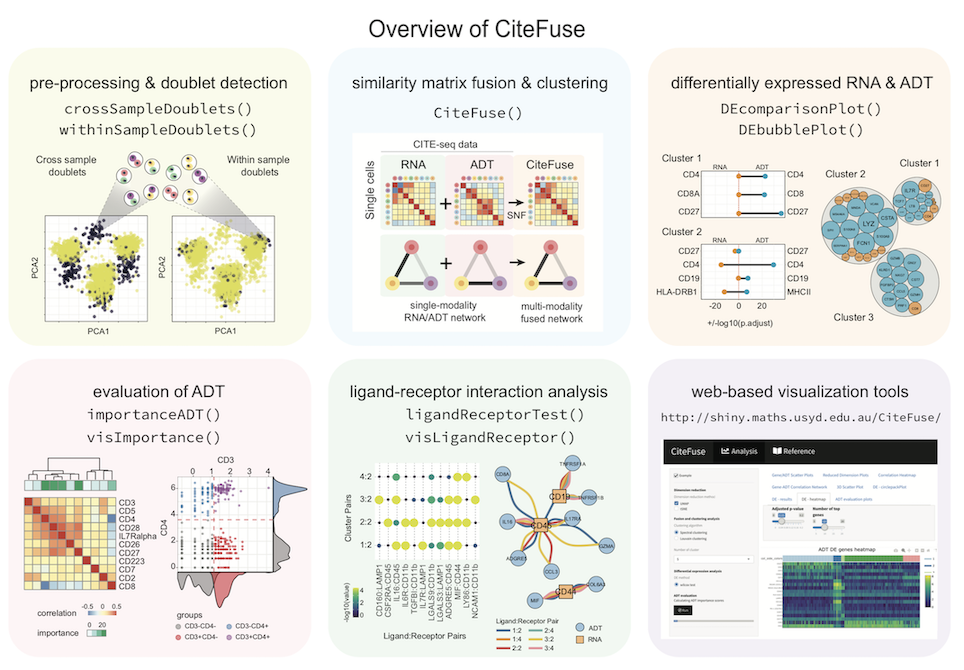
## Citation
Hani Jieun Kim, Yingxin Lin, Thomas A Geddes, Jean Yee Hwa Yang, Pengyi Yang, CiteFuse enables multi-modal analysis of CITE-seq data, _Bioinformatics_, Volume 36, Issue 14, 15 July 2020, Pages 4137–4143, [https://doi.org/10.1093/bioinformatics/btaa282](https://doi.org/10.1093/bioinformatics/btaa282)