Ecosyste.ms: Awesome
An open API service indexing awesome lists of open source software.
https://github.com/s-andrews/redotable
A dotplot application for DNA/RNA sequence
https://github.com/s-andrews/redotable
desktop-application genome-sequencing sequence-alignment sequence-to-sequence visualization
Last synced: 4 months ago
JSON representation
A dotplot application for DNA/RNA sequence
- Host: GitHub
- URL: https://github.com/s-andrews/redotable
- Owner: s-andrews
- License: gpl-3.0
- Created: 2018-08-01T08:00:37.000Z (almost 6 years ago)
- Default Branch: master
- Last Pushed: 2022-11-28T16:10:56.000Z (over 1 year ago)
- Last Synced: 2024-01-17T03:18:23.250Z (6 months ago)
- Topics: desktop-application, genome-sequencing, sequence-alignment, sequence-to-sequence, visualization
- Language: Java
- Size: 388 KB
- Stars: 10
- Watchers: 3
- Forks: 1
- Open Issues: 5
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Lists
- repo-5916-awesome-genome-visualization - Redotable - genome-visualization/redotable.png) (Dotplot)
- awesome-genome-visualization - Redotable
README

Re-dot-able is a dot-plotting application allowing for the visual comparisons of two sets of sequences. It can work interactively, functioning as a desktop application, or can be run as a command line tool.
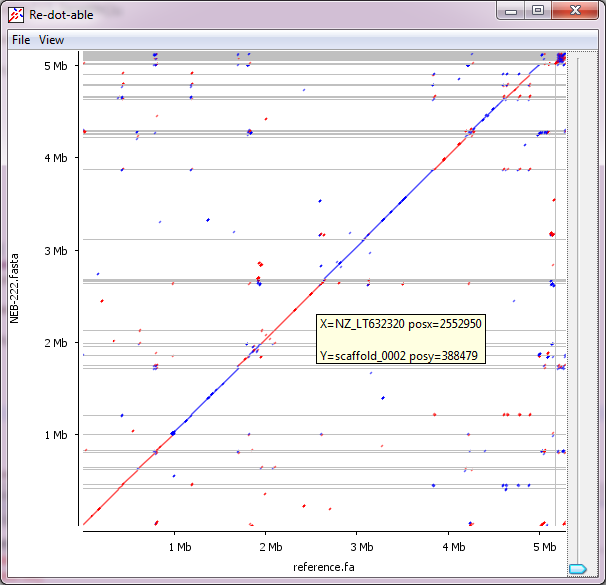
We developed this initially to allow for the comparison of assembled bacterial sequences with reference genomes, but it can be applied to any comparison which involves large sequences.
Installation
============
You can download binary distributions of redotable from the project site at:
www.bioinformatics.babraham.ac.uk/projects/redotable/
Windows
-------
You will need to install the Oracle Java Runtime Environemnt from java.com
Once you have this installed you can download the redotable zip file from the [project web site](https://www.bioinformatics.babraham.ac.uk/projects/redotable/), and uncompress it somewhere on your machine.
To start the program just double click on re-dot-able.exe.
OSX
---
To allow redotable to run on OSX you will need to install the Oracle Java Development Kit which you should be able to get from [here](http://www.oracle.com/technetwork/java/javase/downloads/jdk8-downloads-2133151.html)
Once that is installed you can download the redotable DMG file from the [project web page](https://www.bioinformatics.babraham.ac.uk/projects/redotable/). If you double click on this it will open as a virtual disk and you will see the redotable application icon. To install the program simply drag this icon out of the redotable virtual disk to a location on your computer (for example your Applications folder). You can then run the program by double clicking on the application.
Linux
-----
You will need a full java runtime environment installed on the machine you want to run redotable on. Some distributions will install this by default, others install a cut-down version which can't run graphical applications.
To install a full JRE you can use:
**Ubuntu / Debian** `sudo apt install default-jre`
**CentOS / Redhat** `sudo yum install java-1.8.0-openjdk`
Once this is done you can download and uncompress the windows/linux zip file from the [project web page](https://www.bioinformatics.babraham.ac.uk/projects/redotable/).
You can then launch the program by using the `redotable` launch script in the root folder of the installation.
If you want to add this script to a folder in your path so you can launch the program from anywhere on your system then you will need to do this by creating a symlink to the launch script, **not** by copying it.
For example if you installed redotable into `/opt/redotable`, then to put the program into your path you would do:
`sudo ln -s /opt/redotable/redotable /usr/local/bin/`