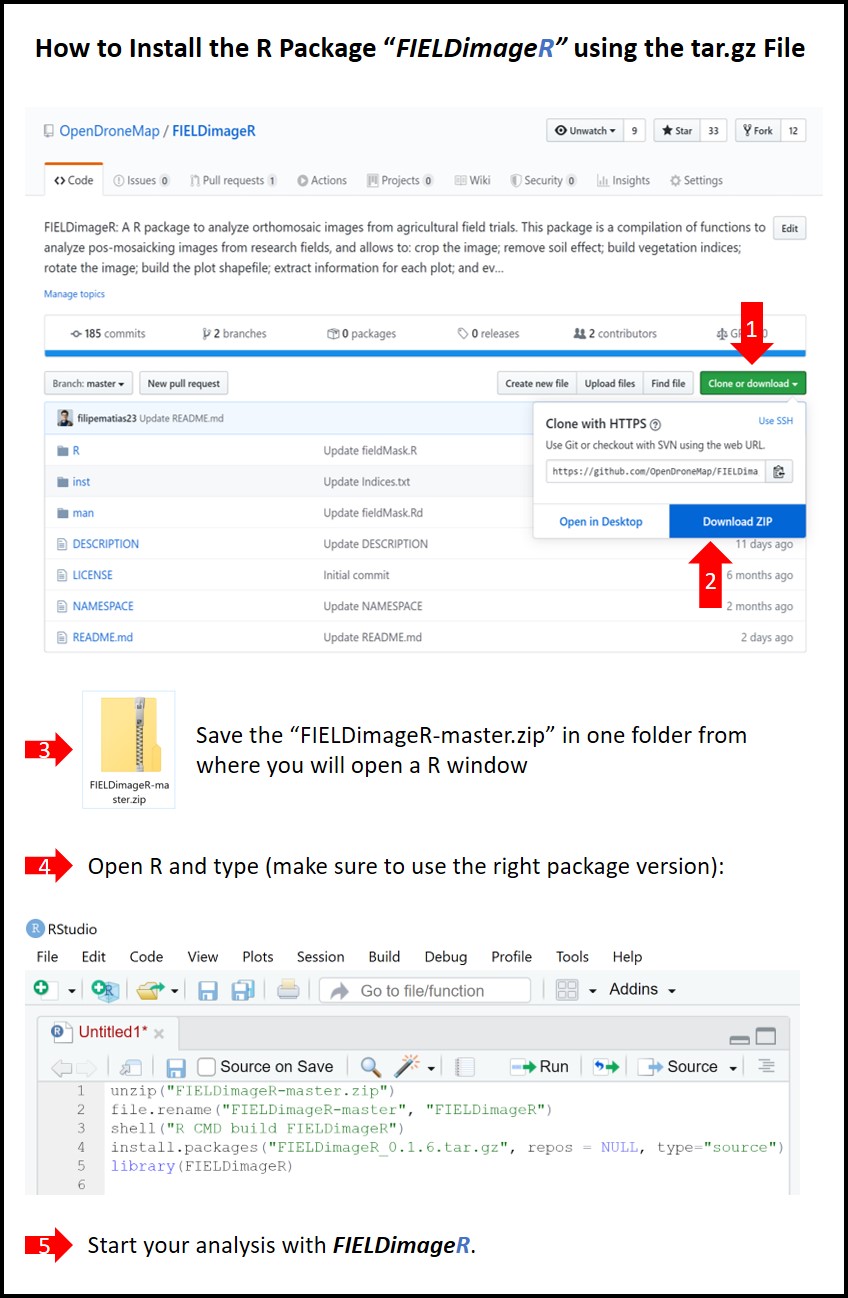
https://github.com/OpenDroneMap/FIELDimageR
FIELDimageR: A R package to analyze orthomosaic images from agricultural field trials. This package is a compilation of functions to analyze pos-mosaicking images from research fields, and allows to: crop the image; remove soil effect; build vegetation indices; rotate the image; build the plot shapefile; extract information for each plot; and evaluate stand count, canopy percentage, and plant height.
https://github.com/OpenDroneMap/FIELDimageR
Last synced: about 2 months ago
JSON representation
FIELDimageR: A R package to analyze orthomosaic images from agricultural field trials. This package is a compilation of functions to analyze pos-mosaicking images from research fields, and allows to: crop the image; remove soil effect; build vegetation indices; rotate the image; build the plot shapefile; extract information for each plot; and evaluate stand count, canopy percentage, and plant height.
- Host: GitHub
- URL: https://github.com/OpenDroneMap/FIELDimageR
- Owner: OpenDroneMap
- License: gpl-2.0
- Created: 2019-11-01T19:36:13.000Z (over 5 years ago)
- Default Branch: master
- Last Pushed: 2024-05-24T18:52:48.000Z (about 1 year ago)
- Last Synced: 2025-05-02T17:03:12.916Z (about 2 months ago)
- Language: R
- Homepage:
- Size: 4.78 MB
- Stars: 253
- Watchers: 40
- Forks: 92
- Open Issues: 28
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
- open-sustainable-technology - FIELDimageR - A Tool to Analyze Images From Agricultural Field Trials and Lab in R. (Consumption / Agriculture and Nutrition)
README
## [FIELDimageR](https://github.com/OpenDroneMap/FIELDimageR): A Tool to Analyze Images From Agricultural Field Trials and Lab in [R](https://www.r-project.org).
> This package is a compilation of functions to analyze orthomosaic images from research fields. To prepare the image it first allows to crop the image, remove soil and weeds and rotate the image. The package also builds a plot shapefile in order to extract information for each plot to evaluate different wavelengths, vegetation indices, stand count, canopy percentage, and plant height.

---------------------------------------------
## Resources
[Installation](#instal)
[1. First steps](#p1)
[2. Loading mosaics and visualizing](#p2)
[3. Removing soil using vegetation indices](#p3)
[4. Building the plot shapefile](#p5)
[5. Building vegetation indices](#p6)
[6. Counting the number of objects (e.g. plants, seeds, etc)](#p7)
[7. Evaluating the object area percentage (e.g. canopy)](#p8)
[8. Extracting data from field images](#p9)
[9. Estimating plant height (e.g., biomass) and creating interpolated mosaics based on sampled points](#p10)
[10. Distance between plants, objects length, and removing objects (plot, cloud, weed, etc.)](#p11)
[11. Resolution and computing time](#p12)
[12. Crop growth cycle](#p13)
[13. Multispectral and Hyperspectral images](#p14)
[14. Building shapefile with polygons (field blocks, pest damage, soil differences, etc)](#p15)
[15. Making plots](#p16)
[16. Saving output files](#p17)
[Orthomosaic using the open source software OpenDroneMap](#p18)
[Parallel and loop to evaluate multiple images (e.g. images from roots, leaves, ears, seed, damaged area, etc.)](#p19)
[Quick tips (memory limits, splitting shapefile, using shapefile from other software, etc)](#p20)
[Contact](#p21)
---------------------------------------------
### Installation
> If desired, one can [build](#Instal_with_docker) a [rocker/rstudio](https://hub.docker.com/r/rocker/rstudio) based [Docker](https://www.docker.com/) image with all the requirements already installed by using the [Dockerfile](https://github.com/OpenDroneMap/FIELDimageR/blob/master/Dockerfile) in this repository.
**With RStudio**
> First of all, install [R](https://www.r-project.org/) and [RStudio](https://rstudio.com/).
> Then, in order to install R/FIELDimageR from GitHub [GitHub repository](https://github.com/OpenDroneMap/FIELDimageR), you need to install the following packages in R. For Windows users who have an R version higher than 4.0, you need to install RTools, tutorial [RTools For Windows](https://cran.r-project.org/bin/windows/Rtools/).
> Now install R/FIELDimageR using the `install_github` function from [devtools](https://github.com/hadley/devtools) package. If necessary, use the argument [*type="source"*](https://www.rdocumentation.org/packages/ghit/versions/0.2.18/topics/install_github).
```r
# You may need other packages first:
install.packages(c('terra','mapview','sf','stars','caret','mapedit','devtools',"dplyr","fields","leafem","leafsync","lwgeom","BiocManager","git2r","exactextractr"))
BiocManager::install("EBImage")
# FIELDimageR:
devtools::install_github("OpenDroneMap/FIELDimageR")
devtools::install_github("filipematias23/FIELDimageR.Extra")
```
> If the method above doesn't work, use the next lines by downloading the FIELDimageR-master.zip file
```r
setwd("~/FIELDimageR-master.zip") # ~ is the path from where you saved the file.zip
unzip("FIELDimageR-master.zip")
file.rename("FIELDimageR-master", "FIELDimageR")
shell("R CMD build FIELDimageR") # or system("R CMD build FIELDimageR")
install.packages("FIELDimageR_0.6.0.tar.gz", repos = NULL, type="source") # Make sure to use the right version (e.g. 0.6.0)
```
**With Docker**
> When building the Docker image you will need the [Dockerfile](https://github.com/OpenDroneMap/FIELDimageR/blob/master/Dockerfile) in this repository available on the local machine.
> Another requirement is that Docker is [installed](https://docs.docker.com/get-docker/) on the machine as well.
>
> Open a terminal window and at the command prompt enter the following command to [build](https://docs.docker.com/engine/reference/commandline/build/) the Docker image:
```bash
docker build -t fieldimager -f ./Dockerfile ./
```
> The different command line parameters are as follows:
>* `docker` is the Docker command itself
>* `build` tells Docker that we want to build an image
>* `-t` indicates that we will be specifying then tag (name) of the created image
>* `fieldimager` is the tag (name) of the image and can be any acceptable name; this needs to immediately follow the `-t` parameter
>* `-f` indicates that we will be providing the name of the Dockerfile containing the instructions for building the image
>* `./Dockerfile` is the full path to the Dockerfile containing the instructions (in this case, it's in the current folder); this needs to immediately follow the `-f` parameter
>* `./` specifies that Docker build should use the current folder as needed (required by Docker build)
>
> The container includes a copy of RStudio Server and `tidyverse` (see [rocker/tidyverse](https://hub.docker.com/r/rocker/tidyverse/)). Alternatively, you can substitute `rocker/tidyverse` in the first line of `Dockerfile` with `rocker/rstudio` for a RStudio environment without the `tidyverse` package.
>
> Once the docker image is built, you use the [Docker run](https://docs.docker.com/engine/reference/run/) command to access the image using the suggested [rocker/rstudio](https://hub.docker.com/r/rocker/rstudio/) command:
```bash
docker run --rm -p 8787:8787 -e PASSWORD=yourpasswordhere fieldimager
```
> Open a web browser window and enter `http://localhost:8787` to access the running container.
> To log into the instance use the username and password of `rstudio` and `yourpasswordhere`.
#### If you are using anaconda and Linux
> To install this package on Linux and anaconda it is necessary to use a series of commands before the recommendations
* Install Xorg dependencies for the plot system (on conda shell)
```
conda install -c conda-forge xorg-libx11
```
* Install the BiocManager package manager
```r
install.packages("BiocManager")
```
* Use the BiocManager to install the EBIMAGE package
```r
BiocManager::install("EBImage")
```
* If there is an error in the fftw3 library header (fftw3.h) install the dependency (on conda shell)
```
conda install -c eumetsat fftw3
```
* If there is an error in the dependency doParallel
```r
install.packages ("doParallel")
```
* Continue installation
```r
setwd("~/FIELDimageR-master.zip") # ~ is the path from where you saved the file.zip
unzip("FIELDimageR-master.zip")
file.rename("FIELDimageR-master", "FIELDimageR")
system("R CMD build FIELDimageR") #only system works on linux
install.packages("FIELDimageR_0.6.0.tar.gz", repos = NULL, type="source") # Make sure to use the right version (e.g. 0.6.0)
```
[Menu](#menu)
---------------------------------------------
### Using R/FIELDimageR
#### 1. First steps
> **Taking the first step:**
```r
library(FIELDimageR)
library(FIELDimageR.Extra)
library(raster)
library(terra)
library(mapview)
library(sf)
library(stars)
```
[Menu](#menu)
---------------------------------------------
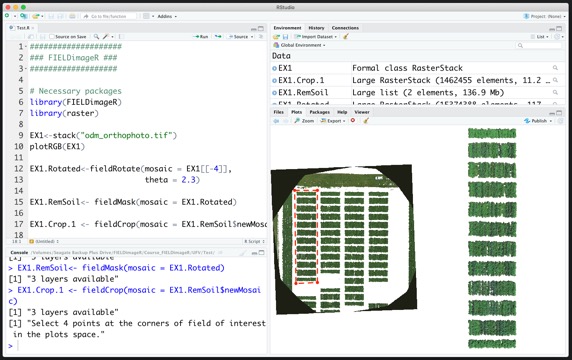
#### 2. Loading mosaics and visualizing
> The following example uses an image available to download here: [EX1_RGB.tif](https://drive.google.com/open?id=1S9MyX12De94swjtDuEXMZKhIIHbXkXKt). If necessary, the image/mosaic size can be reduced around the field boundaries for faster image analysis using the function: **`fieldCrop`**.
```r
# Uploading an example mosaic
EX1 <- rast("EX1_RGB.tif")
# Visualization Option-01 (FIELDimageR.Extra):
fieldView(EX1)
# Visualization Option-02 (raster):
plotRGB(EX1, r = 1, g = 2, b = 3)
# Only if necessary you can crop the mosaic/image to reduce size:
# x11()
# EX1 <- rast(fieldCrop(mosaic = EX1)) # For heavy images (large, high resolution, etc.) please use: fast.plot=T
```

```r
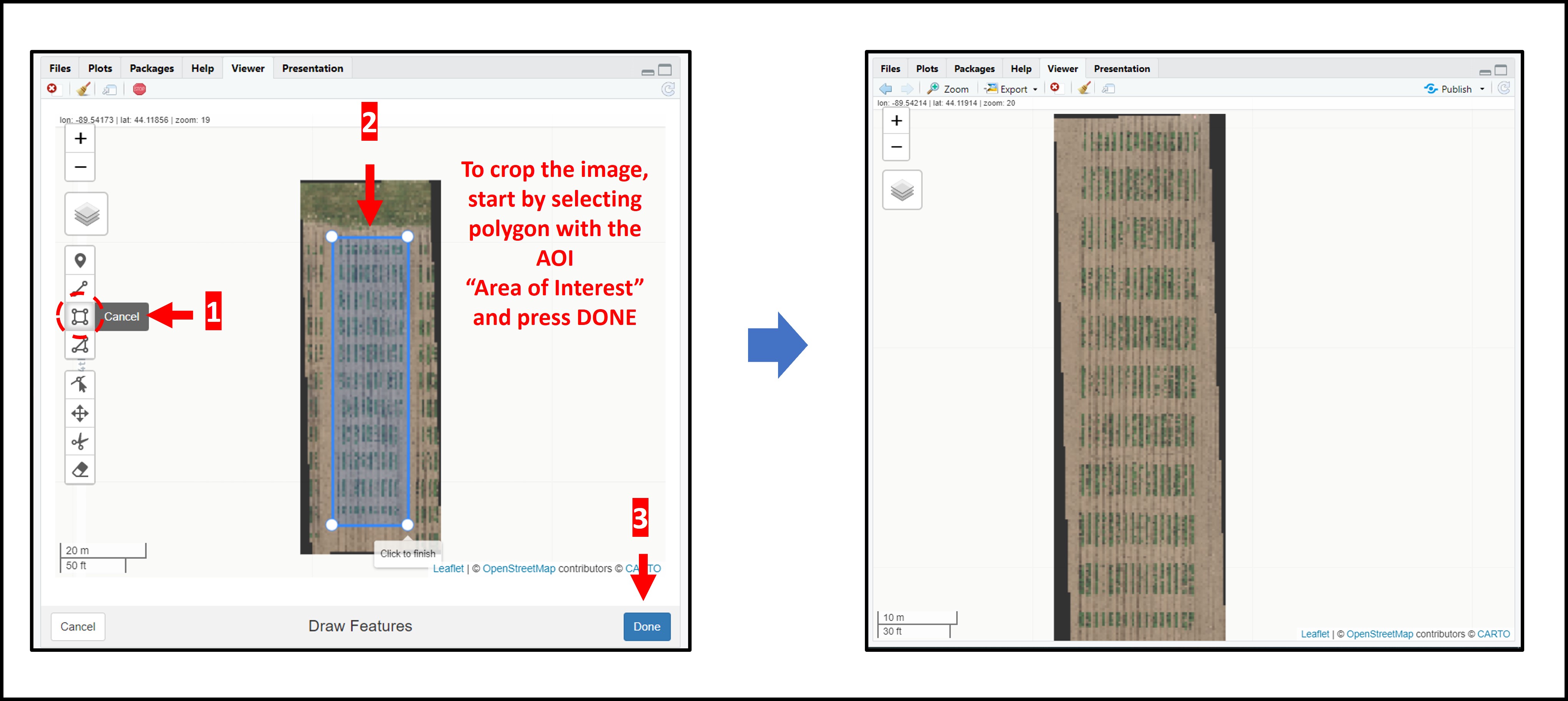
# If necessary you can crop the mosaic/image to reduce size by selecting the aoi (area of interest) using fieldView with editor=TRUE:
aoi<-fieldView(EX1,editor = TRUE)
EX1<-crop(EX1,aoi) # For heavy images (large, high resolution, etc.) please use: fast.plot=T
fieldView(EX1)
```
[Menu](#menu)
---------------------------------------------
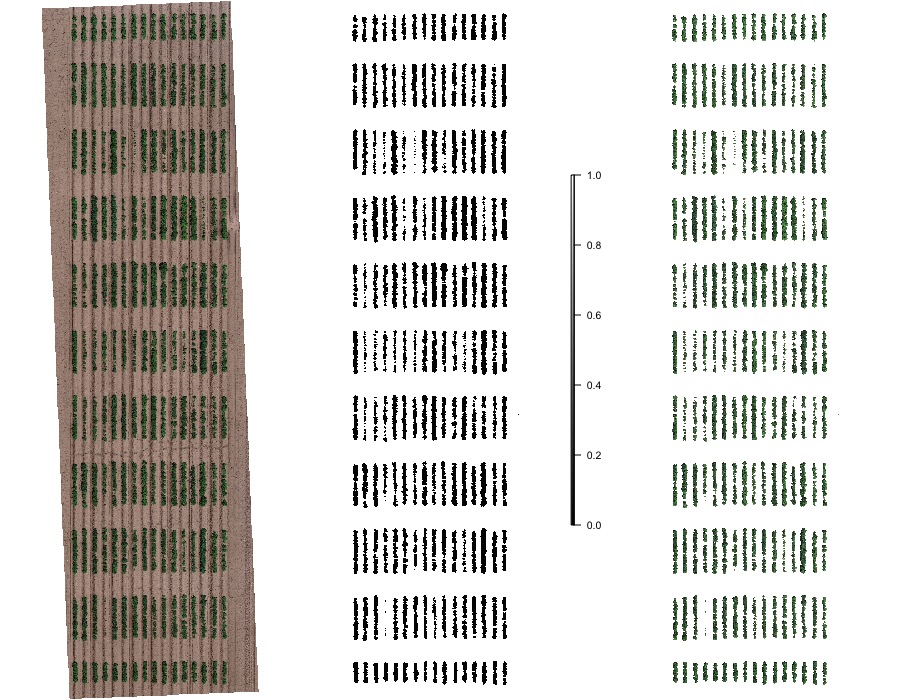
#### 3. Removing soil using vegetation indices
> The presence of soil can introduce bias in the data extracted from the image. Therefore, removing soil from the image is one of the most important steps for image analysis in agricultural science. Function to use: **`fieldMask`**
```r
EX1.RemSoil <- fieldMask(mosaic = EX1, Red = 1, Green = 2, Blue = 3, index = "HUE")
```
[Menu](#menu)
---------------------------------------------
#### 4. Building the plot shape file
> Once the field has reached a correct straight position, the plot shape file can be drawn by selecting at least four points at the corners of the experiment. The number of columns and rows must be informed. At this point the experimental borders can be eliminated, in the example bellow the borders were removed in all the sides. Function to use is from [FIELDimageR.Extra: **`fieldShape_render`**](https://github.com/filipematias23/FIELDimageR.Extra#p3)
```r
EX1.Shape<-fieldShape_render(mosaic = EX1,ncols = 14, nrows = 9)
fieldView(mosaic = EX1,
fieldShape = EX1.Shape,
type = 2,
alpha = 0.2)
```

> **Attention:** The plots are identified in ascending order from *left to right* and *bottom to top* (this is another difference from the **`FIELDimageR::fieldShape`** ) being evenly spaced and distributed inside the selected area independent of alleys.
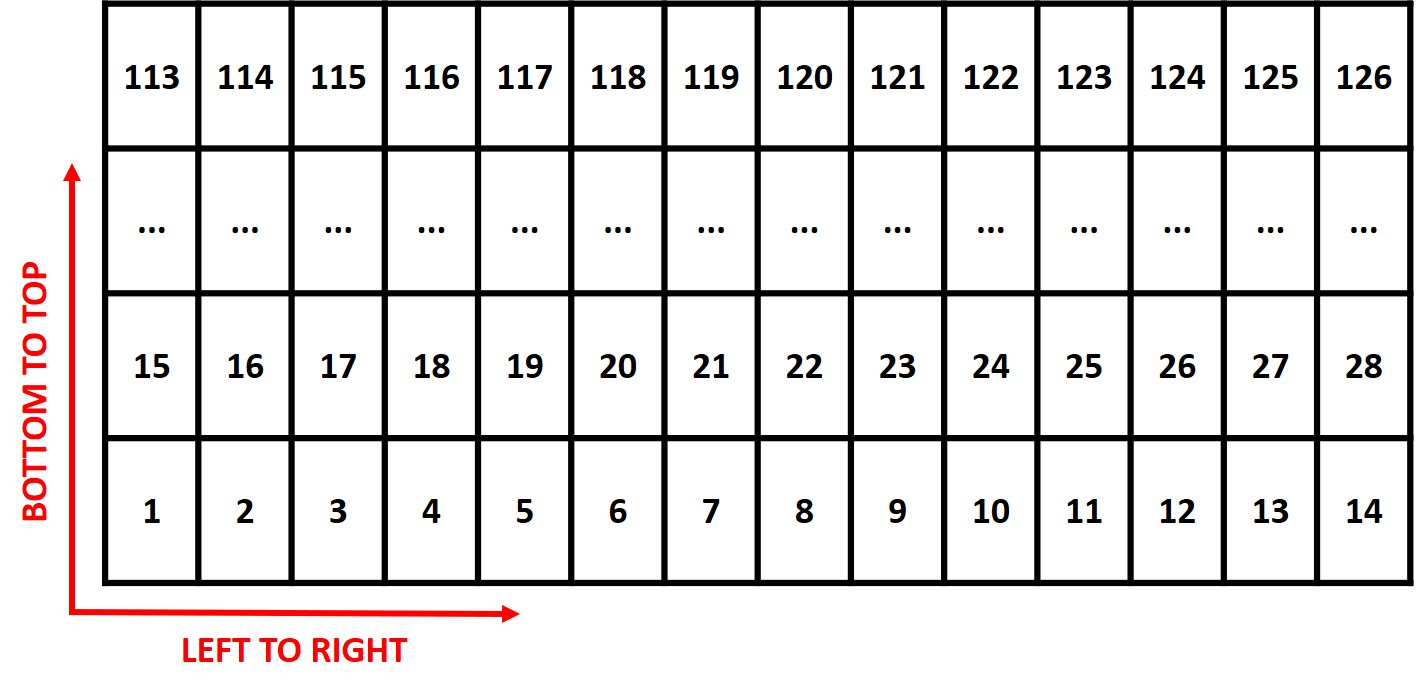
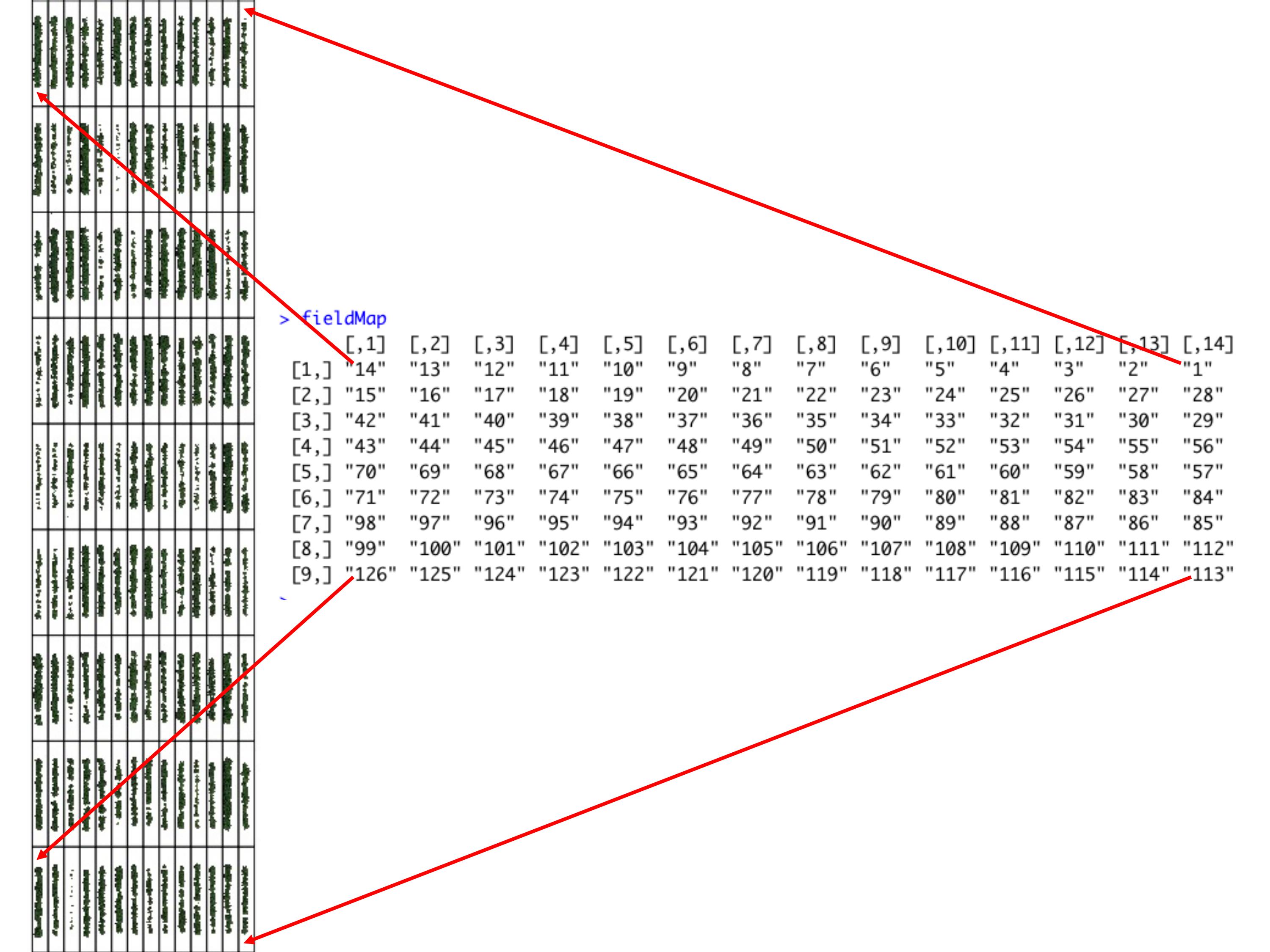
> One matrix can be used to identify the plots position according to the image above. The function **`fieldMap`** can be used to specify the plot *ID* automatic or any other matrix (manually built) also can be used. For instance, the new column **PlotName** will be the new identification. You can download an external table example here: [DataTable.csv](https://drive.google.com/open?id=18YE4dlSY1Czk2nKeHgwd9xBX8Yu6RCl7).
```r
### Field map identification (name for each Plot). 'fieldPlot' argument can be a number or name.
DataTable<-read.csv("DataTable.csv",header = T)
fieldMap<-fieldMap(fieldPlot=DataTable$Plot, fieldColumn=DataTable$Row, fieldRow=DataTable$Range, decreasing=T)
fieldMap
# The new column PlotName is identifying the plots:
EX1.Shape<-fieldShape_render(mosaic = EX1, ncols = 14, nrows = 9, fieldMap = fieldMap, fieldData = DataTable, PlotID = "Plot")
fieldView(mosaic = EX1,
fieldShape = EX1.Shape,
type = 2,
alpha = 0.2)
```
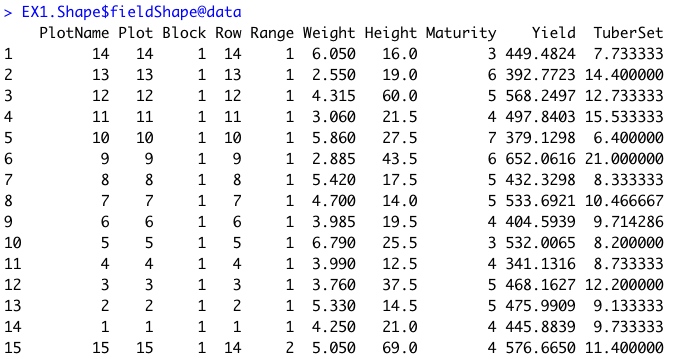
```r
### Joing all information in one "fieldShape_render" file:
EX1.Shape<-fieldShape_render(mosaic = EX1, ncols = 14, nrows = 9, fieldMap = fieldMap, fieldData = DataTable, PlotID = "Plot")
fieldView(mosaic = EX1,
fieldShape = EX1.Shape,
type = 2,
alpha = 0.2)
# The new column PlotName is identifying the plots:
EX1.Shape
```
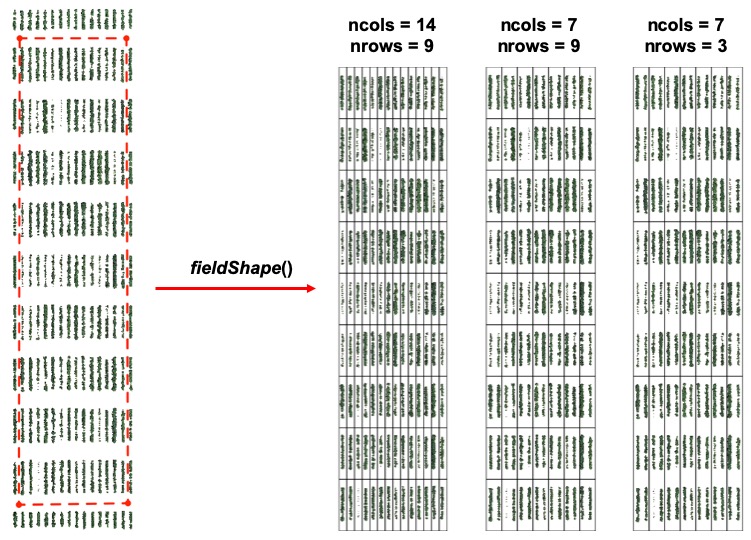
```r
### Different plot dimensions using "fieldShape":
# ncols = 14 and nrows = 9
EX1.Shape.1Line<-fieldShape_render(mosaic = EX1, ncols = 14, nrows = 9)
# ncols = 7 and nrows = 9
EX1.Shape.2lines<-fieldShape_render(mosaic = EX1, ncols = 7, nrows = 9)
# ncols = 7 and nrows = 3
EX1.Shape.6lines<-fieldShape_render(mosaic = EX1, ncols = 7, nrows = 3)
```
[Menu](#menu)
---------------------------------------------
#### 5. Building vegetation indices
> A general number of indices are implemented in *FIELDimageR* using the function **`fieldIndex`**. Also, you can build your own index using the parameter `myIndex`.
```r
# Calculating myIndex = "(Red-Blue)/Green" (not avaliable at 'FIELDimageR')
EX1.Indices<- fieldIndex(mosaic = EX1.RemSoil$newMosaic, Red = 1, Green = 2, Blue = 3,
index = c("NGRDI","BGI"),
myIndex = c("(Red-Blue)/Green"))
# More than one myIndex code: myIndex = c("myIndex1","myIndex2","myIndex3")
EX1.Indices.myIndex<- fieldIndex(mosaic = EX1.RemSoil$newMosaic, Red = 1, Green = 2, Blue = 3,
index = c("NGRDI","BGI"),
myIndex = c("(Red-Blue)/Green","Red/Green","Blue/Green"))
```

> *Sugestion:* This function could also be used to build an index to remove soil or weeds. First it is necessary to identify the threshold to differentiate soil from the plant material. At the example below (B), all values above 0.7 were considered as soil and further removed using **`fieldMask`** (C & D).
```r
EX1.Indices.BGI<- fieldIndex(mosaic = EX1, index = c("BGI"))
dev.off()
hist(EX1.Indices.BGI$BGI) # Image segmentation start from 0.7 (soil and plants)
EX1.BGI<- fieldMask(mosaic = EX1, Red = 1, Green = 2, Blue = 3,
index = "BGI", cropValue = 0.7, cropAbove = T)
#Check if: cropValue=0.8 or cropValue=0.6 works better.
```

[Menu](#menu)
---------------------------------------------
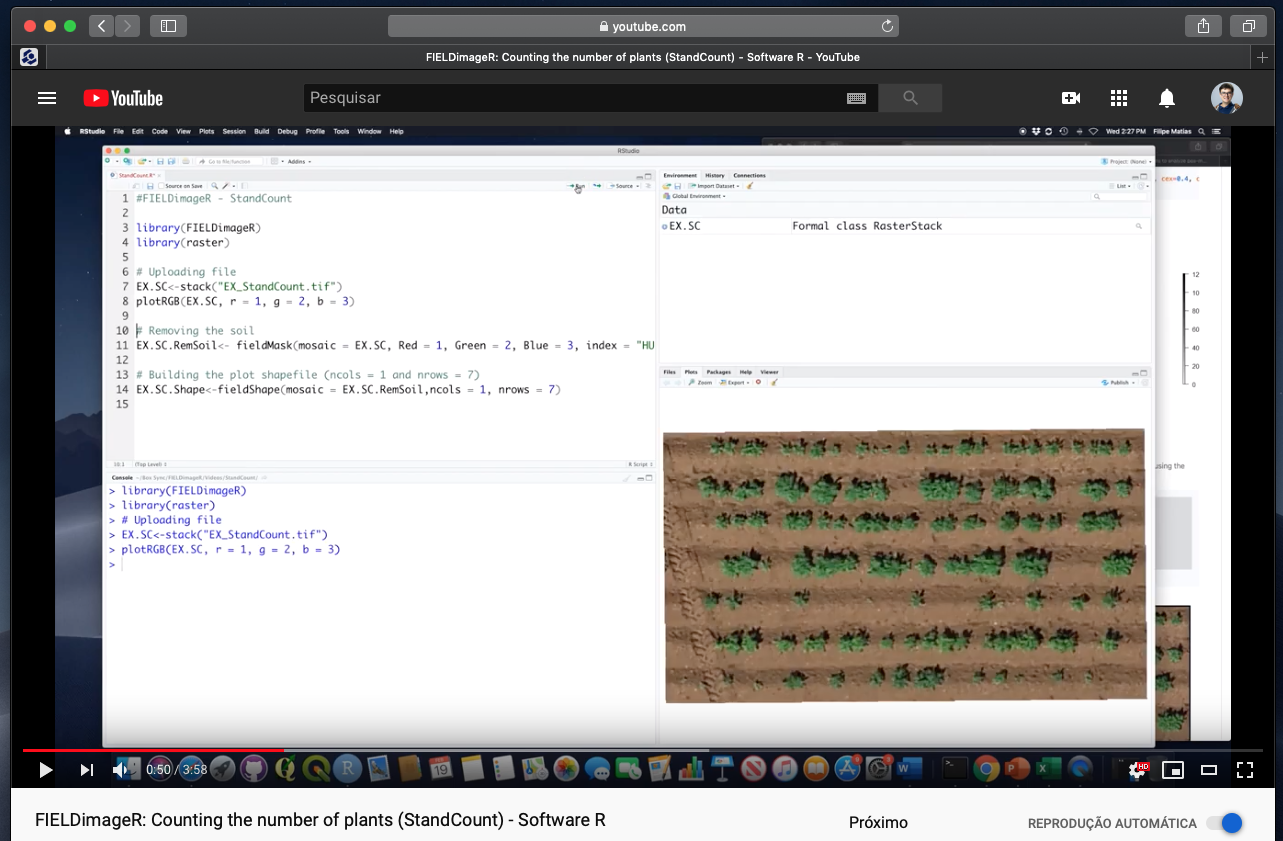
#### 6. Counting the number of objects (e.g. plants, seeds, etc)
> *FIELDimageR* can be used to evaluate stand count during early stages. A good weed control practice should be performed to avoid misidentification inside the plot. The *mask* output from **`fieldMask`** and the *fieldshape* output from **`fieldShape`** must be used. Function to use: **`fieldCount`**.
```r
EX1.SC<-fieldCount(mosaic = EX1.RemSoil$mask,
fieldShape = EX1.Shape,
plot=T,
col="blue")
```
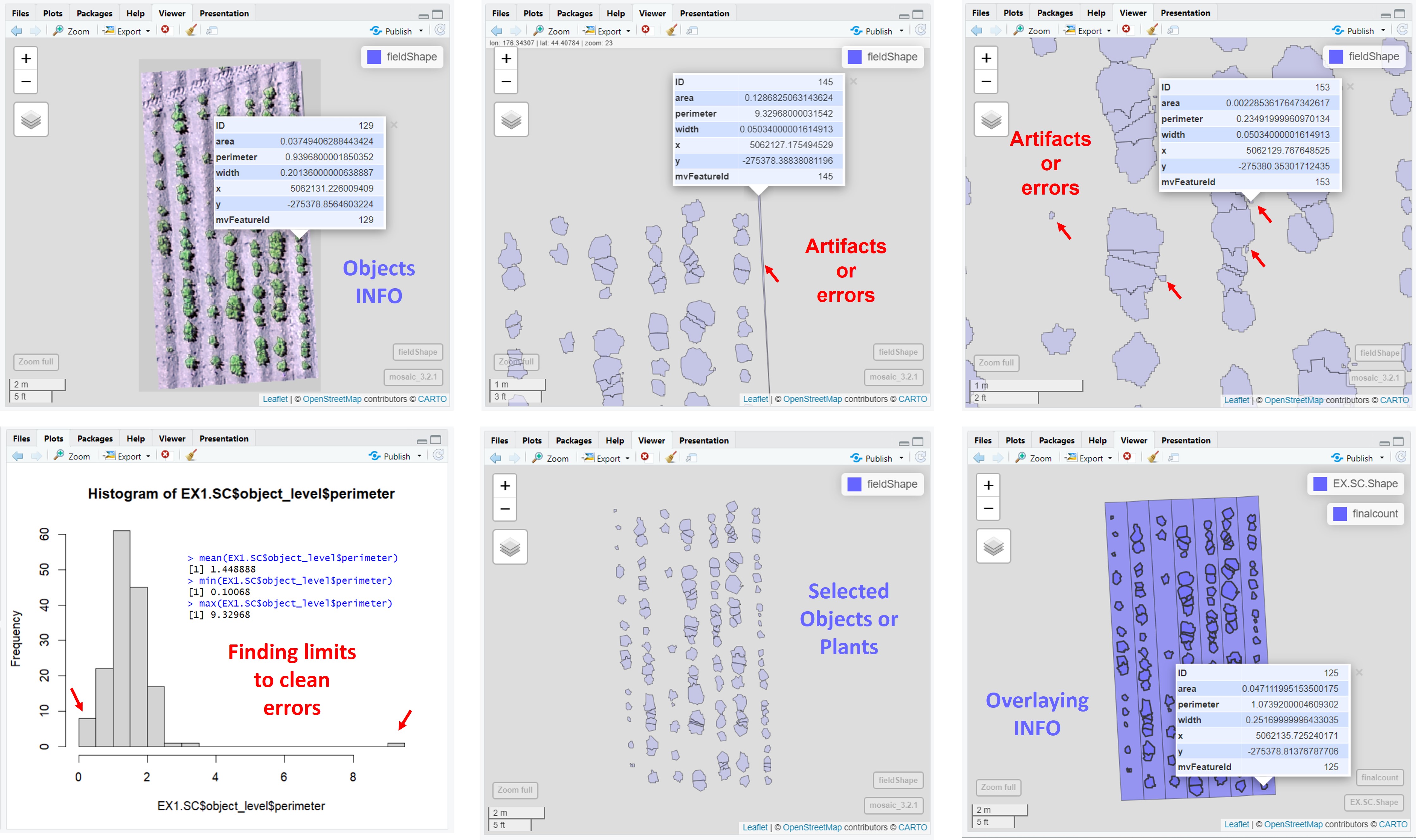
```r
# New shapeFile with objects in the plot. Data per plot in the grid: area, perimeter, count, and mean_width.
EX1.SC$plot_level
fieldView(mosaic = EX1.RemSoil$newMosaic,
fieldShape = EX1.SC$plot_level,
type = 2,
alpha = 0.2)
# New shapeFile of single objects. Data per object (plant, pollen, etc.): area, perimeter, width, x and y position.
EX1.SC$object_level
fieldView(mosaic = EX1.RemSoil$newMosaic,
fieldShape = EX1.SC$object_level,
type = 2,
alpha = 0.5)
```

> To refine stand count, we can further eliminate weeds (small plants) or outlying branches from the output using the parameter *min Size*. The following example uses an image available to download here:[EX_StandCount.tif](https://drive.google.com/open?id=1PzWcIsYQMxgEozR5HHUhT0RKcstMOfSk)
```r
# Uploading file
EX.SC<-rast("EX_StandCount.tif")
plotRGB(EX.SC, r = 1, g = 2, b = 3)
# Removing the soil
EX.SC.RemSoil<- fieldMask(mosaic = EX.SC, Red = 1, Green = 2, Blue = 3, index = "HUE")
# Building the plot shapefile (ncols = 7 and nrows = 1)
EX.SC.Shape<-fieldShape_render(mosaic = EX.SC,
ncols = 7,
nrows = 1)
fieldView(mosaic = EX.SC,
fieldShape = EX.SC.Shape,
type = 2,
alpha = 0.2)
```

```r
### When all shapes are counted: minSize = 0.00
EX1.SC<-fieldCount(mosaic = EX.SC.RemSoil$mask,
fieldShape = EX.SC.Shape,
watershed = 0.05,
plot = T)
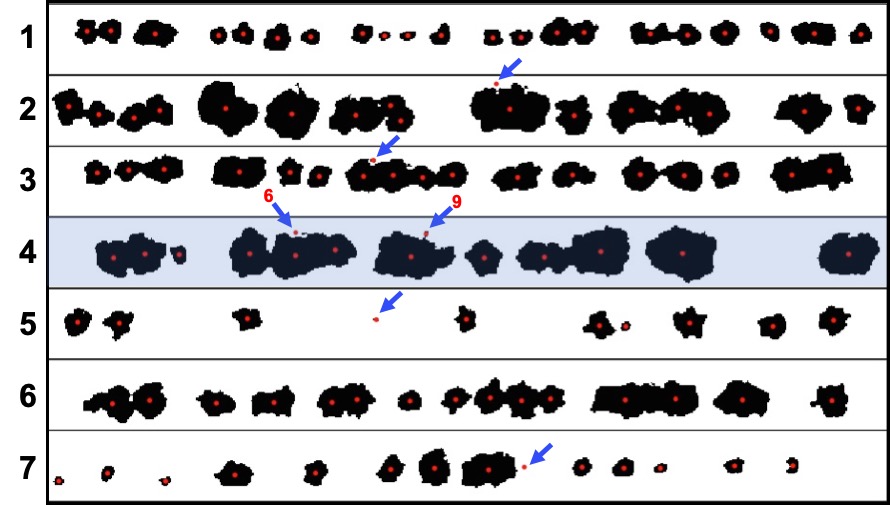
EX1.SC$object_level # Identifies plants and small artifacts
fieldView(mosaic = EX.SC,
fieldShape = EX1.SC$object_level,
type = 2,
alpha = 0.2)
```
```r
### When all shapes with perimeter greater than 0.4 and lower than 4 are selected:
hist(EX1.SC$object_level$perimeter,breaks=30) # Errors= 0.4 < perimeter > 4
max(EX1.SC$object_level$perimeter)
min(EX1.SC$object_level$perimeter)
mean(EX1.SC$object_level$perimeter)
finalcount<-EX1.SC$object_level %>% dplyr::filter(perimeter>0.4 & perimeter<4) # Errors= 0.4 < perimeter > 4
mapview(list(EX.SC.Shape,finalcount))
fieldView(mosaic = EX.SC,
fieldShape = finalcount,
type = 2,
alpha = 0.2)
```
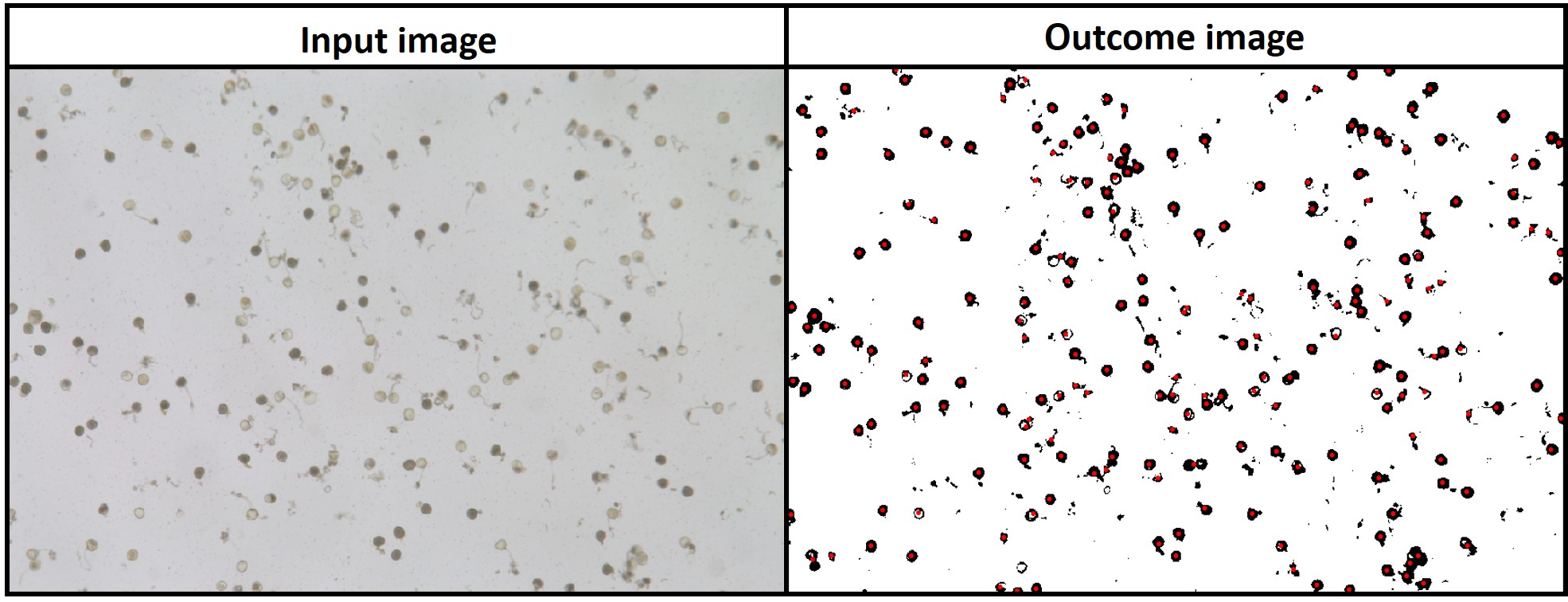
> **`fieldCount`** also can be used to count other objects (e.g. seed, pollen, etc). The example below *FIELDimageR* pipeline was used to count seeds in a corn ear.

> **`fieldMask`** with index BIM (Brightness Index Modified) was used to identify pollen and **`fieldCount`** was used to count total and germinated pollens per sample. [Download EX_Pollen.jpeg](https://drive.google.com/open?id=1Tyr4cEvEBoaWqaHw8UWPW-X1unzEMAfk)
```r
# Uploading image
EX.P<-rast("EX_Pollen.jpeg")
# Reducing image resolution (fast analysis)
EX.P<-aggregate(EX.P,fact=4)
plotRGB(EX.P, r = 1, g = 2, b = 3)
# Using index "BIM" to remove background (above 19)
EX.P<-imgLAB(EX.P)
EX.P.R1<- fieldMask(mosaic = EX.P,index = "BIM", cropValue = 19, cropAbove = T)
fieldView(EX.P.R1$newMosaic)
# Counting all pollens area>1000 (all sample)
EX.P.Total<-fieldCount(mosaic = EX.P.R1$mask, plot=T)
library(leafsync)
m1<-fieldView(EX.P)
m2<-mapview(EX.P.Total)
sync(m1,m2)
hist(EX.P.Total$area,breaks=50)
pollen_count<-EX.P.Total %>% dplyr::filter(area>1000)
m2<-mapview(pollen_count)
sync(m1,m2)
# Using index "BIM" to identify germinated pollen grain (removing values above 16)
EX.P.R2<- fieldMask(mosaic = EX.P, index = "BIM", cropValue = 16, cropAbove = T)
m2<-fieldView(EX.P.R2$newMosaic)
sync(m1,m2)
# Counting all germinated pollen
EX.P.Ger<-fieldCount(mosaic = EX.P.R2$mask, plot=T)
m2<-mapview(EX.P.Ger)
sync(m1,m2)
```
[Menu](#menu)
---------------------------------------------
#### 7. Evaluating the object area percentage (e.g. canopy)
> *FIELDimageR* can also be used to evaluate the canopy percentage per plot. The *mask* output from **`fieldMask`** and the *fieldshape* output from **`fieldShape`** must be used. Function to use: **`fieldArea`**. The parameter *n.core* is used to accelerate the canopy extraction (parallel).
```r
EX1.Canopy<-fieldArea(mosaic = EX1.RemSoil$newMosaic, fieldShape = EX1.Shape)
EX1.Canopy
```

> **`fieldArea`** also can be associated with the function **`fieldMask`** to evaluate the proportion of seed colors in Glass Gems Maize. An important step is to choose the right contrasting background to differentiate samples. The same approach can be used to evaluate the percentage of disease/pests’ damage, land degradation, deforestation, etc.

[Menu](#menu)
---------------------------------------------
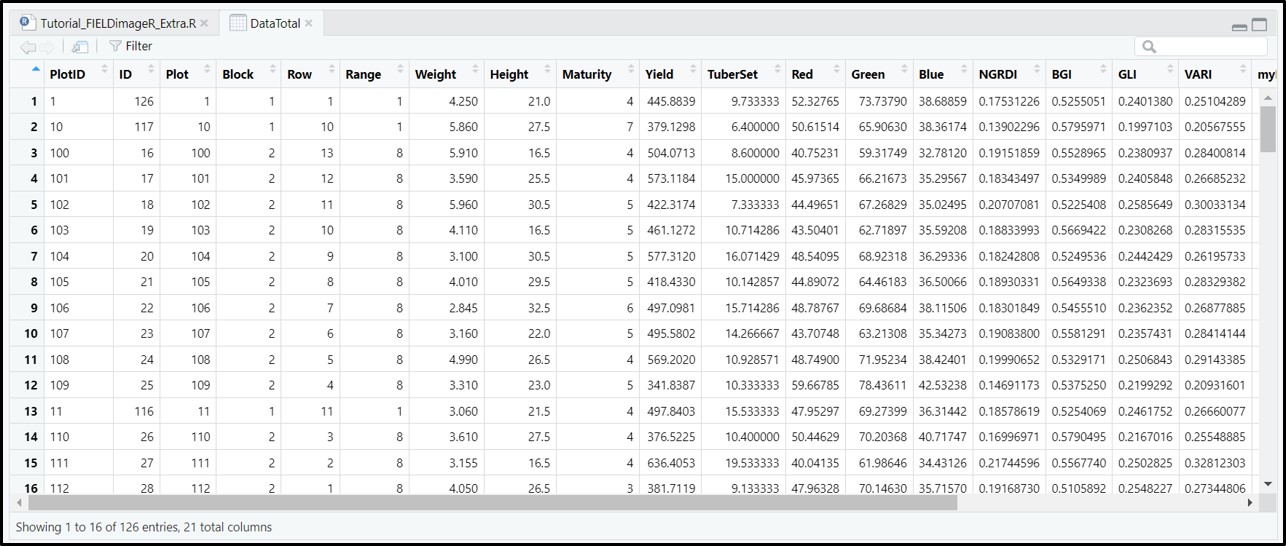
#### 8. Extracting data from field images
> The function *extract* from **[terra](https://CRAN.R-project.org/package=terra)** is adapted for agricultural field experiments through function [**`FIELDimageR.Extra::fieldInfo_extra`**](https://github.com/filipematias23/FIELDimageR.Extra#p6).
```r
EX1.Info<- fieldInfo_extra(mosaic = EX1.Indices,fieldShape = EX1.Shape)
EX1.Info
```
[Menu](#menu)
---------------------------------------------
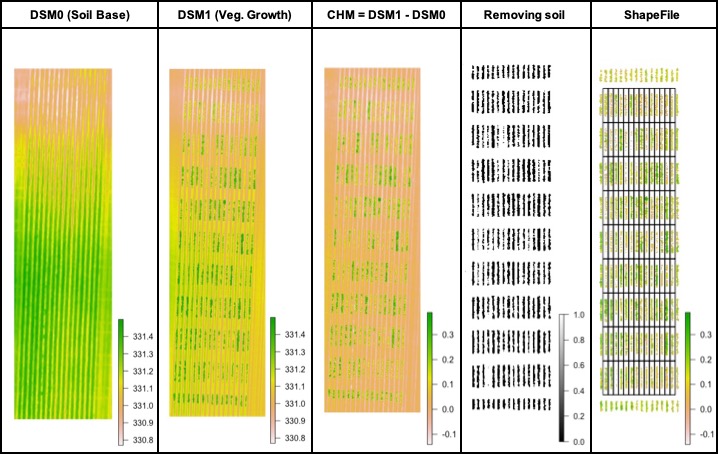
#### 9. Estimating plant height and biomass
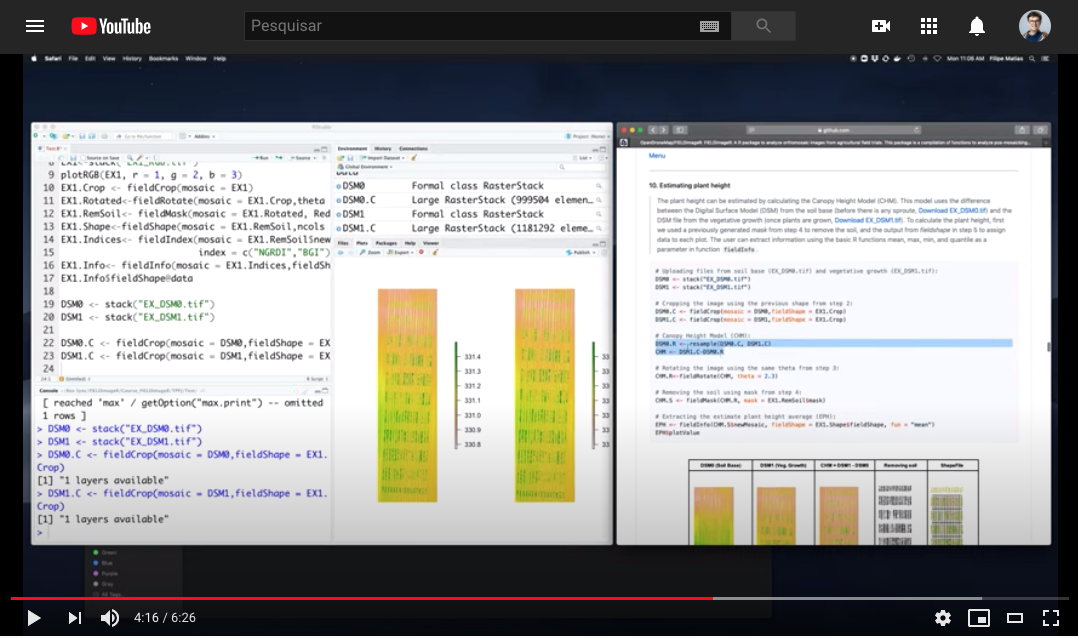
> The plant height can be estimated by calculating the Canopy Height Model (CHM) and biomass by calculating Canopy Volume Model (CVM). This model uses the difference between the Digital Surface Model (DSM) from the soil base (before there is any sproute, [Download EX_DSM0.tif](https://drive.google.com/open?id=1lrq-5T6x_GrbkCtpDSDiX1ldvSwEBFX-)) and the DSM file from the vegetative growth (once plants are grown, [Download EX_DSM1.tif](https://drive.google.com/open?id=1q_H4Ef1f1yQJOPtkVMJfcb2SvHcxJ3ya)). To calculate CHM and CVM we used the function **`fieldHeight`**, where CVM=cellSize(CHM)*CHM. The next step is removing the soil effect with **`fieldMask`** and then use **`fieldInfo_extra`** to extract information as mean, max, min, sum, and quantile.
```r
# Uploading files from soil base (EX_DSM0.tif) and vegetative growth (EX_DSM1.tif):
DSM0 <- rast("EX_DSM0.tif")
DSM1 <- rast("EX_DSM1.tif")
# Canopy Height Model (CHM) and Canopy Volume Model (CVM):
CHVM<-fieldHeight(DSM0,DSM1)
plot(CHVM)
# Removing the soil using mask from step 4:
CHVM <- fieldMask(CHVM, mask = EX1.RemSoil$mask)
# Extracting the estimate plant height average (PlantHeight):
EX1.Shape<- fieldInfo_extra(mosaic = CHVM$newMosaic$height,
fieldShape = EX1.Shape,
fun=mean)
colnames(EX1.Shape)[dim(EX1.Shape)[2]-1]<-"PlantHeight" # Changing trait name!
# Extracting the estimate plant height at 10% and 90% of quantile:
probs = c(0.1,0.9)
EPH.Extract<-extract(x = CHVM$newMosaic$height, y = EX1.Shape,quantile, probs = probs, na.rm=TRUE)
EX1.Shape<-merge(EX1.Shape,EPH.Extract,by="ID")
colnames(EX1.Shape)[c(dim(EX1.Shape)[2]-2,
dim(EX1.Shape)[2]-1)]<-c("PH.10","PH.90") # Changing trait name!
# Extracting plant volume or digital biomass (PlantBiomass):
EX1.Shape<- fieldInfo_extra(mosaic = CHVM$newMosaic$volume,
fieldShape = EX1.Shape,
fun=sum)
colnames(EX1.Shape)[dim(EX1.Shape)[2]-1]<-"PlantBiomass" # Changing trait name!
# Data Visualization:
m1<-fieldView(EX1)
m2<-fieldView(mosaic = CHVM$newMosaic$height,
fieldShape = EX1.Shape,
type = 2,
alpha = 0.5)
library(leafsync)
sync(m1,m2)
```
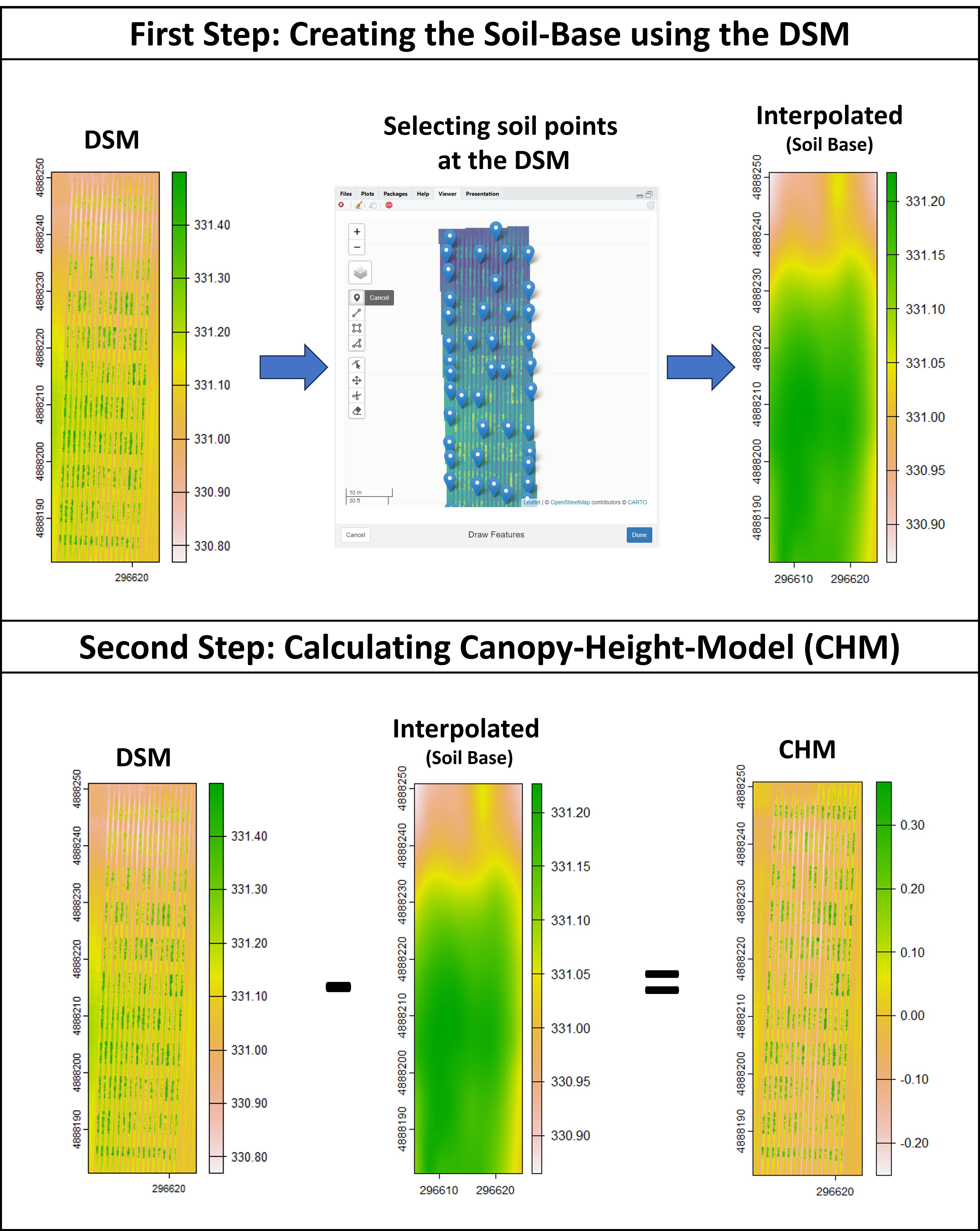
### Creating interpolated mosaics based on sampled points
> Attention: In case the user has only one flight (e.g., one DSM) the function **`fieldInterpolate`** can be used to creat the soil-reference based on sampled points at the DSM1.
```r
# Creating the soil reference based on the DSM1:
DSM1 <- rast("EX_DSM1.tif")
# Selecting points from soil:
points<-fieldView(DSM1,editor = T)
# Creating the interpolated soil-base (DSM0.New):
DSM0.New<-fieldInterpolate(DSM1,points)
plot(DSM0.New)
# New Canopy-Height-Model (CHM):
CHM<-DSM1-DSM0.New
plot(CHM)
```
[Menu](#menu)
---------------------------------------------
#### 10. Distance between plants, objects length, and removing objects (plot, cloud, weed, etc.)
> The function **`fieldObject`** can be used to take relative measurement from objects (e.g., area, "x" distance, "y" distance, number, extent, etc.) in the entire mosaic or per plot using the fieldShape file. Download example here: [EX_Obj.jpg](https://drive.google.com/file/d/1F189a1LKA9_l0Xn8hdBOshL5eC_fQLPs/view?usp=sharing).
```r
# Uploading file (EX_Obj.tif)
EX.Obj <- rast("EX_Obj.jpg")
plotRGB(EX.Obj)
EX.Obj <- aggregate(EX.Obj,4)
# Removing the background
EX.Obj<-imgLAB(EX.Obj)
EX.Obj.M<- fieldMask(mosaic = EX.Obj, index = "BGI",cropValue = 0.7,cropAbove = T)
# Taking measurements:
EX.Obj.D<-fieldCount(mosaic = EX.Obj.M$mask,
plot = T)
fieldView(EX.Obj,EX.Obj.D)
plotRGB(EX.Obj)
plot(EX.Obj.D$geometry, add=T, border="red")
plot(EX.Obj.D[1,], add=T, col="yellow")
EX.Obj.I<- fieldIndex(mosaic = EX.Obj,index = c("SI","BGI","BI"))
EX.Obj.Data<-fieldInfo_extra(mosaic = EX.Obj.I[[c("SI","BGI","BI")]],
fieldShape = EX.Obj.D)
plot(EX.Obj.Data)
# Data visualization:
library(reshape2)
Data.Obj1<-melt(EX.Obj.Data,measure.vars = c("area","perimeter","width","SI","BGI","BI"))
library(ggplot2)
ggplot(Data.Obj1, aes(x=value, fill=variable)) +
geom_histogram(aes(y=..density..), colour="black")+
geom_density(alpha=.2)+
facet_wrap(~variable, scales = "free")
```

> The function **`fieldCrop`** can be used to remove objects from the field image. For instance, the parameter *remove=TRUE* and *nPoint* should be used to select the object boundaries to be removed. [Download EX_RemObj.tif](https://drive.google.com/open?id=1wfxSQANRrPOvJWwNZ6UU0UjXwzKfkKH0)).
```r
# Uploading file (EX_RemObj.tif)
EX.RemObj <- rast("EX_RemObj.tif")
# Selecting the object boundaries to be removed (nPoint = 10)
EX.RemObj.Crop <- fieldCrop(mosaic = EX.RemObj, remove = T, nPoint = 10) # Selecting the plant in plot 13
# Removing the soil
EX.RemObj.RemSoil<- fieldMask(mosaic = EX.RemObj.Crop,index = "HUE")
# Building the plot shapefile (ncols = 8 and nrows = 4)
EX.RemObj.Shape<-fieldShape_render(mosaic = EX.RemObj.RemSoil,ncols = 8, nrows = 4)
# Building indice (NGRDI)
EX.RemObj.Indices<- fieldIndex(mosaic = EX.RemObj.RemSoil$newMosaic,index = c("NGRDI"))
# Extracting data (NGRDI)
EX.RemObj.Info<- fieldInfo_extra(mosaic = EX.RemObj.Indices$NGRDI,
fieldShape = EX.RemObj.Shape)
# Comparing plots values (the plant in plot 13 was removed and its value must be lower than plot 12 and 14)
EX.RemObj.Info[c(12,13,14),]
```

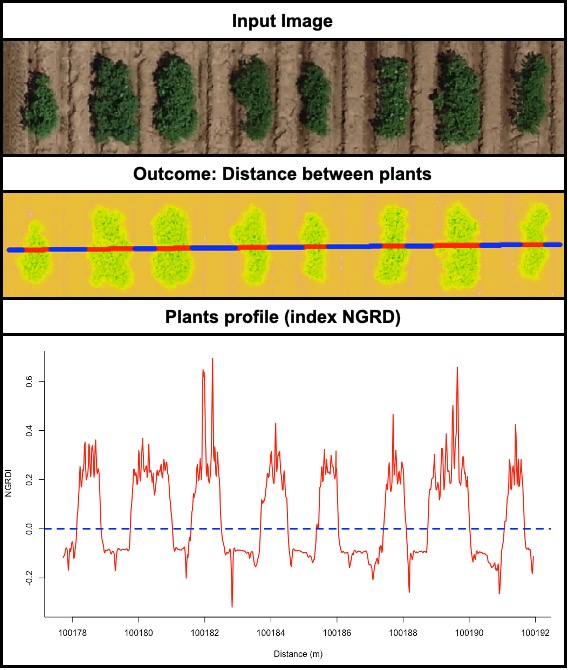
> The function **`fieldDraw`** can be used to draw lines or polygons in the field image. This function allows to extract information of specific positions in the field (x, y, value). Also, this function can be used to evaluate distances between objects (for example: distance between plants in a line) or either objects length (for example: seed length, threes diameter, etc.). Let's use the image above to evaluate the distance between potato plots and later to extract NGRDI values from a line with soil and vegetation to observe the profile in a distance plot.
```r
# Uploading file (EX_RemObj.tif)
EX.Dist <- rast("EX_RemObj.tif")
# vegetation indices
EX.Dist.Ind <- fieldIndex(mosaic = EX.Dist,index = c("NGRDI"))
# Removing the soil
EX.Dist.RemSoil <- fieldMask(mosaic = EX.Dist.Ind)
# Evaluating distance between plants. Remember to press ESC when finished to draw the line.
EX.Dist.Draw <- fieldDraw(mosaic = EX.Dist.RemSoil$mask,
dist = T,
value = 1, # Use the number to identify the object to be measured (1 for space and 0 for plants)
distSel = 0.4)
EX.Dist.Draw$drawDist # Distance between plants
# Making plots
plot(EX.Dist.Ind$NGRDI)
points(EX.Dist.Draw$drawData$x,EX.Dist.Draw$drawData$y, col="red",pch=16,cex=0.7)
points(EX.Dist.Draw$drawSegments$x,EX.Dist.Draw$drawSegments$y, col="blue",pch=16,cex=0.7)
lines(EX.Dist.Draw$drawDist[1,c("x1","x2")],EX.Dist.Draw$drawDist[1,c("y1","y2")], col="green",lwd=5)
# Evaluating an specific layer profile (e.g. NGRDI)
EX.Dist.Draw.2 <- fieldDraw(mosaic = EX.Dist.Ind,
ndraw = 4, # Making 4 lines (press ESC to conclude each line)
lwd = 5)
EX.Data<-EX.Dist.Draw.2$Draw1$drawData
dev.off()
plot(x = EX.Data$x, y = EX.Data$NGRDI, type="l", col="red",lwd=2,xlab="Distance (m)", ylab="NGRDI")
abline(h=0.0,col="blue", lty=2, lwd=3)
# Making polygons and extracting data per cell
EX.Dist.Draw.3 <- fieldDraw(mosaic = EX.Dist.Ind,
line = F, # Making 2 polygons (press ESC to conclude each polygon)
ndraw = 2,
lwd = 5)
plotRGB(EX.Dist.RemSoil$newMosaic)
plot(EX.Dist.Draw.3$Draw1$drawObject, col="red",add=T)
```
[Menu](#menu)
---------------------------------------------
#### 11. Resolution and computing time
> The influence of image resolution was evaluated at different steps of the FIELDimageR pipeline. For this propose, the resolution of image *EX1_RGB_HighResolution.tif* [Download](https://drive.google.com/open?id=1elZe2jfq4bQSZM8cFAS4q7fRrnXbSBgH) was reduced using the function **raster::aggregate** in order to simulate different flown altitudes Above Ground Surface (AGS). The parameter *fact* was used to modify the original image resolution (0.4x0.4 cm with 15m AGS) to: first, **fact=2** to reduce the original image to 0.8x0.8 cm (simulating 30m AGS), and **fact=4** to reduce the original image to 1.6x1.6 (simulating 60m AGS). The steps (*i*) removing soil, (*ii*) building vegetation index (BGI), and (*iii*) getting information were evaluated using the function **system.time** output *elapsed* (R base).
```r
### Image and resolution decrease
RES_1<-rast("EX1_RGB_HighResolution.tif")
RES_2<-aggregate(RES_1, fact=2)
RES_4<-aggregate(RES_1, fact=4)
res(RES_1)
res(RES_2)
res(RES_4)
par(mfrow=c(1,3))
plotRGB(RES_1)
plotRGB(RES_2)
plotRGB(RES_4)
### Removing Soil
system.time({RES_1_S <- fieldMask(RES_1,index="HUE")})
system.time({RES_2_S <- fieldMask(RES_2,index="HUE")})
system.time({RES_4_S <- fieldMask(RES_4,index="HUE")})
### Indices
system.time({RES_1_I <- fieldIndex(RES_1_S$newMosaic,index=c("BGI"))})
system.time({RES_2_I <- fieldIndex(RES_2_S$newMosaic,index=c("BGI"))})
system.time({RES_4_I <- fieldIndex(RES_4_S$newMosaic,index=c("BGI"))})
### Get Information (1 Band)
system.time({RES_1_Info <- fieldInfo_extra(RES_1_I$BGI,fieldShape = EX1.Shape)})
system.time({RES_2_Info <- fieldInfo_extra(RES_2_I$BGI,fieldShape = EX1.Shape)})
system.time({RES_4_Info <- fieldInfo_extra(RES_4_I$BGI,fieldShape = EX1.Shape)})
### Get Information (3 Bands)
system.time({RES_1_Info2 <- fieldInfo_extra(RES_1_I[[c(1,2,3)]],fieldShape = EX1.Shape)})
system.time({RES_2_Info2 <- fieldInfo_extra(RES_2_I[[c(1,2,3)]],fieldShape = EX1.Shape)})
system.time({RES_4_Info2 <- fieldInfo_extra(RES_4_I[[c(1,2,3)]],fieldShape = EX1.Shape)})
### Correlation
DataBGI <- data.frame(R1=RES_1_Info$BGI,
R2=RES_2_Info$BGI,
R4=RES_4_Info$BGI)
DataBlue <- data.frame(R1=RES_1_Info2$Blue,
R2=RES_2_Info2$Blue,
R4=RES_4_Info2$Blue)
DataGreen <- data.frame(R1=RES_1_Info2$Green,
R2=RES_2_Info2$Green,
R4=RES_4_Info2$Green)
DataRed <- data.frame(R1=RES_1_Info2$Red,
R2=RES_2_Info2$Red,
R4=RES_4_Info2$Red)
cor(DataBGI)
cor(DataBlue)
cor(DataGreen)
cor(DataRed)
```
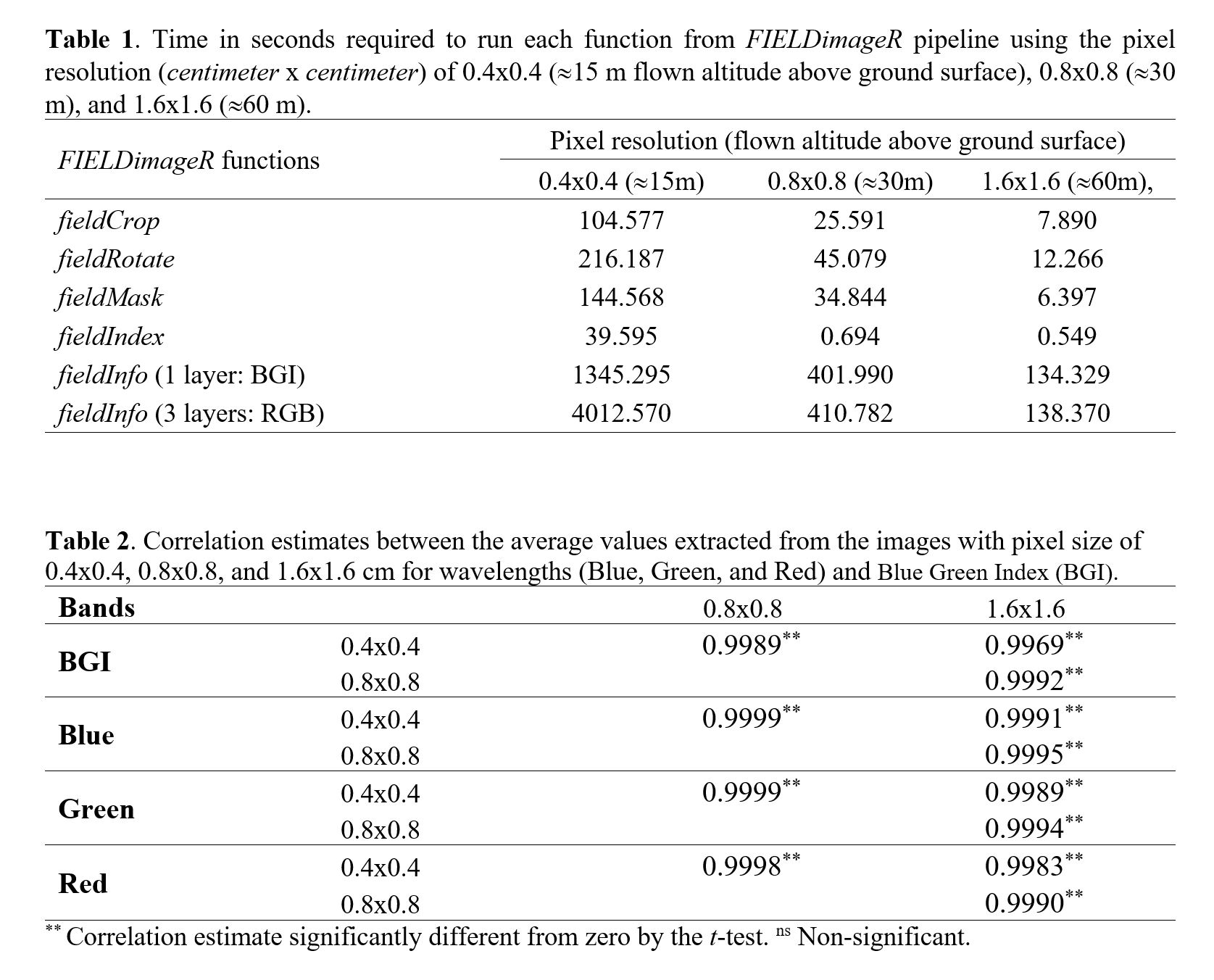
> The time to run one function using the image with pixel size of 0.4x0.4 cm can be 10 (**`fieldInfo`**) to 70 times (**`fieldIndex`**) slower than the image with pixel size of 1.6x1.6 cm (Table 1). The computing time to extract BGI index (one layer) with 0.4x0.4 cm was ~23 min whereas only ~7 min with the 0.8x0.8 cm image, and ~2 min using the 1.6x1.6 cm image. the time to extract the RGB information (three layers) was ~2.3 min for the 1.6x1.6 cm image and ~66 min for the 0.4x0.4 cm image. It is important to highlight that the resolution did not affect the plots mean, it has a correlation >99% between 0.4x0.4 cm and 1.6x1.6 (Table 2). High resolution images showed to require higher computational performance, memory, and storage space. We experienced that during the image collection in the field a low altitudes flight needs more batteries and a much greater number of pictures, and consequently longer preprocessing images steps to build ortho-mosaics.
[Menu](#menu)
---------------------------------------------
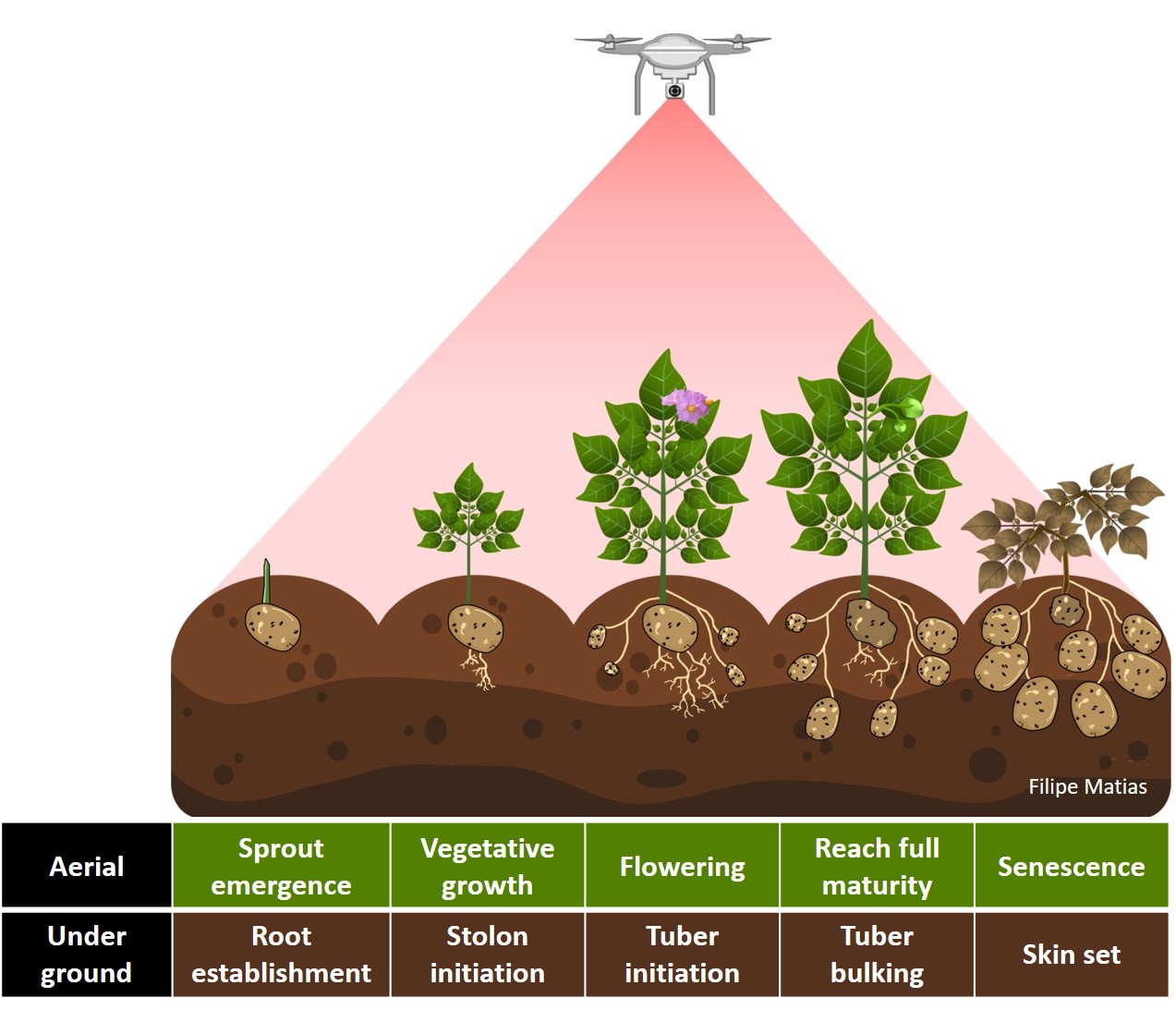
#### 12. Crop growth cycle
> The same rotation theta from step 3, mask from step 4, and plot shape file from step 5, can be used to evaluate mosaics from other stages in the crop growth cycle. Here you can download specific images from flowering and senecense stages in potatoes. ([**Flowering: EX2_RGB.tif**](https://drive.google.com/open?id=1B1HrIYUVqSpKdDN8E8VudpI8jT8MYbWY) and [**Senescence: EX3_RGB.tif**](https://drive.google.com/open?id=15GpLy669mICpkorbUk1M9vqfSUMHbdc5))
```r
# Uploading Flowering (EX2_RGB.tif) and Senescence (EX3_RGB.tif) files:
EX2 <- rast("EX2_RGB.tif")
EX3 <- rast("EX3_RGB.tif")
# Cropping the image using the previous shape from step 2:
EX2.Crop <- fieldCrop(mosaic = EX2,fieldShape = EX1.Crop, plot = T)
EX3.Crop <- fieldCrop(mosaic = EX3,fieldShape = EX1.Crop, plot = T)
# Rotating the image using the same theta from step 3:
EX2.Rotated<-fieldRotate(EX2.Crop,theta = 2.3, plot = T)
EX3.Rotated<-fieldRotate(EX3.Crop,theta = 2.3, plot = T)
# Removing the soil using index and mask from step 4:
EX2.RemSoil<-fieldMask(EX2.Rotated,index="HUE",cropValue=0,cropAbove=T,plot=T)
EX3.RS<-fieldMask(EX3.Rotated,index="HUE",cropValue=0,cropAbove=T,plot=T) # Removing soil at senescence stage
EX3.RemSoil<-fieldMask(EX3.RS$newMosaic,mask = EX2.RemSoil$mask ,cropValue=0,cropAbove=T,plot=T) # Removing weeds from senescence stage with flowering mask
# Building indices
EX2.Indices <- fieldIndex(EX2.RemSoil$newMosaic,Red=1,Green=2,Blue=3,
index = c("NGRDI","BGI"), myIndex = c("(Red-Blue)/Green"))
EX3.Indices <- fieldIndex(EX3.RemSoil$newMosaic,Red=1,Green=2,Blue=3,
index = c("NGRDI","BGI"), myIndex = c("(Red-Blue)/Green"))
# Extracting data using the same fieldShape file from step 5:
EX2.Info<- fieldInfo_extra(mosaic = EX2.Indices$myIndex,fieldShape = EX1.Shape)
EX3.Info<- fieldInfo_extra(mosaic = EX3.Indices$myIndex,fieldShape = EX1.Shape)
Data.Cycle<-data.frame(EX1=EX1.Info$myIndex,
EX2=EX2.Info$myIndex,
EX3=EX3.Info$myIndex)
Data.Cycle
```


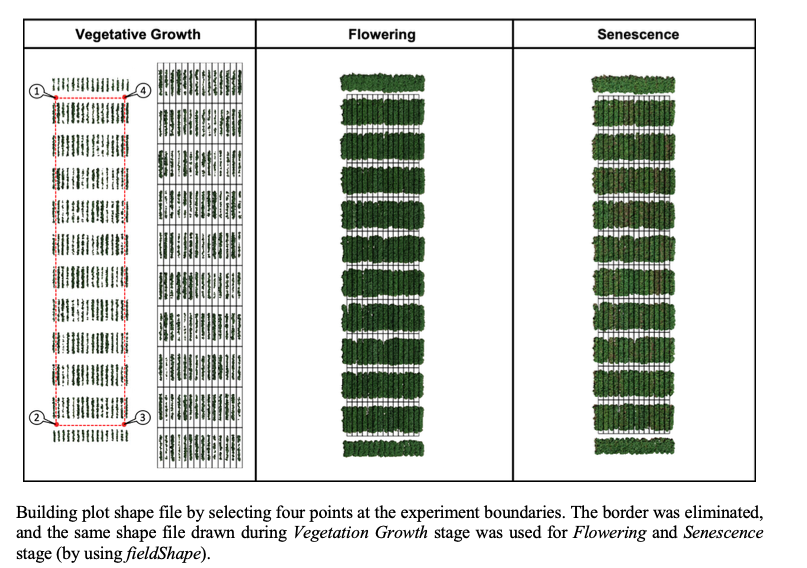
[Menu](#menu)
---------------------------------------------
#### 13. Multispectral and Hyperspectral images
> **`FIELDimageR`** can be used to analyze multispectral and hyperspectral images. The same rotation theta, mask, and plot shape file used to analyze RGB mosaic above can be used to analyze multispectral or hyperspectral mosaic from the same field.
**Multispectral:** You can dowload a multispectral example here: [**EX1_5Band.tif**](https://drive.google.com/open?id=1vYb3l41yHgzBiscXm_va8HInQsJR1d5Y)
```r
#####################
### Multispectral ###
#####################
# Uploading multispectral mosaic:
EX1.5b <- rast("EX1_5Band.tif")
# Cropping the image using the previous shape from step 2:
EX1.5b.Crop <- fieldCrop(mosaic = EX1.5b,fieldShape = EX1.Crop, plot = T)
# Rotating the image using the same theta from step 3:
EX1.5b.Rotated<-fieldRotate(EX1.5b.Crop,theta = 2.3, plot = T)
# Removing the soil using index and mask from step 4:
EX1.5b.RemSoil<-fieldMask(EX1.5b.Rotated,Red=1,Green=2,Blue=3,index="HUE",cropValue=0,cropAbove=T,plot=T)
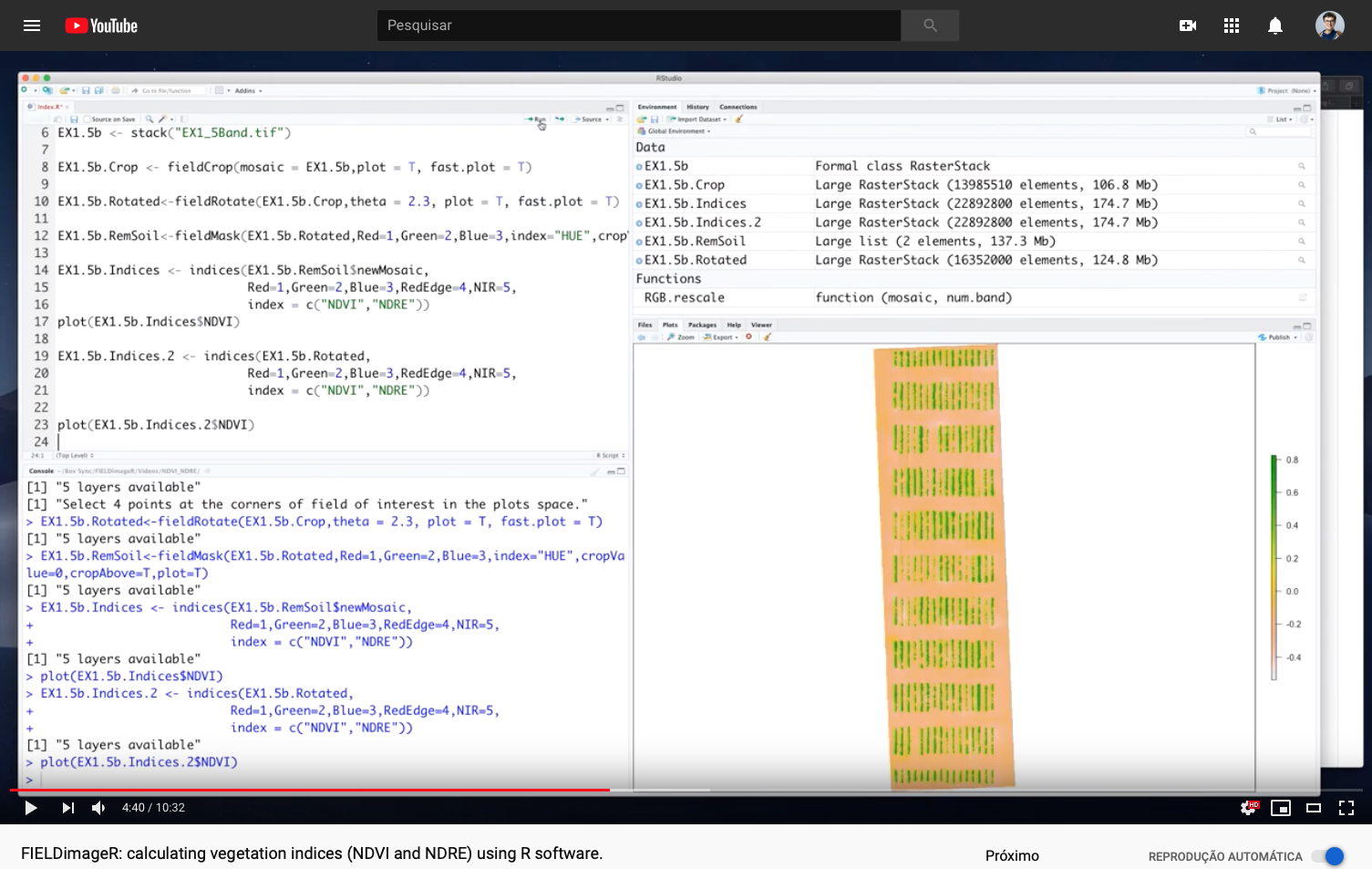
# Building indices (NDVI and NDRE)
EX1.5b.Indices <- fieldIndex(EX1.5b.RemSoil$newMosaic,Red=1,Green=2,Blue=3,RedEdge=4,NIR=5,
index = c("NDVI","NDRE"))
# Extracting data using the same fieldShape file from step 5:
EX1.5b.Info<- fieldInfo_extra(mosaic = EX1.5b.Indices$NDVI,fieldShape = EX1.Shape)
```
**Hyperspectral:** in the following example you should download the hyperspectral tif file ([**EX_HYP.tif**](https://drive.google.com/file/d/1oOKUJ4sA1skyU7NU7ozTQZrT7_s24luW/view?usp=sharing)), the 474 wavelength names ([**namesHYP.csv**](https://drive.google.com/file/d/1n-3JX0ho1rMSOiGrrYmBKHkeF4hyHQw1/view?usp=sharing)), and the field map data ([**DataHYP.csv**](https://drive.google.com/file/d/1u78jb9BlA45kVaXUu2RKM6_KJjXIpJXq/view?usp=sharing)).
```r
#####################
### Hyperspectral ###
#####################
# Uploading hyperspectral file with 474 bands (EX_HYP.tif)
EX.HYP<-rast("EX_HYP.tif")
# Wavelengths (namesHYP.csv)
NamesHYP<-as.character(read.csv("namesHYP.csv")$NameHYP)
# Building RGB image
R<-EX.HYP[[78]] # 651nm (Red)
G<-EX.HYP[[46]] # 549nm (Green)
B<-EX.HYP[[15]] # 450nm (Blue)
RGB<-rast(c(R,G,B))
plotRGB(RGB, stretch="lin")
# Removing soil using RGB (index NGRDI)
RGB.S<-fieldMask(RGB,index="NGRDI",cropValue = 0.0, cropAbove = F)
# Data frame with field information to make the Map
Data<-read.csv("DataHYP.csv")
Map<-fieldMap(fieldPlot = as.character(Data$Plot),fieldRow = as.character(Data$Range),fieldColumn = as.character(Data$Row),decreasing = T)
# Building plot shapefile using RGB as base
plotFile<-fieldShape_render(RGB.S,ncols = 14, nrows = 14, fieldMap = Map,fieldData = Data, PlotID = "Plot")
# Removing soil using the RGB mask
EX.HYP.S<-fieldMask(EX.HYP,mask = RGB.S$mask, plot = F)
# Extracting data (474 bands)
EX.HYP.I<-fieldInfo_extra(EX.HYP.S$newMosaic,fieldShape = plotFile)
# Saving the new csv with hyperspectral information per plot
DataHYP<-EX.HYP.I[,-dim(EX.HYP.I)[2]]
colnames(DataHYP)<-c(colnames(DataHYP)[1:9],NamesHYP)
write.csv(DataHYP,"DataHypNew.csv",col.names = T,row.names = F)
###############
### Graphic ###
###############
dev.off()
DataHYP1<-EX.HYP.I$plotValue[,-1]
plot(x=as.numeric(NamesHYP),y=as.numeric(DataHYP1[1,]),type = "l",xlab = "Wavelength (nm)",ylab = "Reflectance", col="black",lwd=2,cex.lab=1.2)
for(i in 2:dim(DataHYP1)[2]){
lines(x=as.numeric(NamesHYP),y=as.numeric(DataHYP1[i,]),type = "l",col=i,lwd=2)
}
abline(v=445,col="blue",lwd=2,lty=2)
abline(v=545,col="green",lwd=2,lty=2)
abline(v=650,col="red",lwd=2,lty=2)
abline(v=720,col="red",lwd=2,lty=3)
abline(v=840,col="red",lwd=2,lty=4)
legend(list(x = 2000,y = 0.5),c("Blue (445nm)","Green (545nm)","Red (650nm)","RedEdge (720nm)","NIR (840nm)"),
col =c("blue","green","red","red","red"),lty=c(2,2,2,3,4),box.lty=0)
```
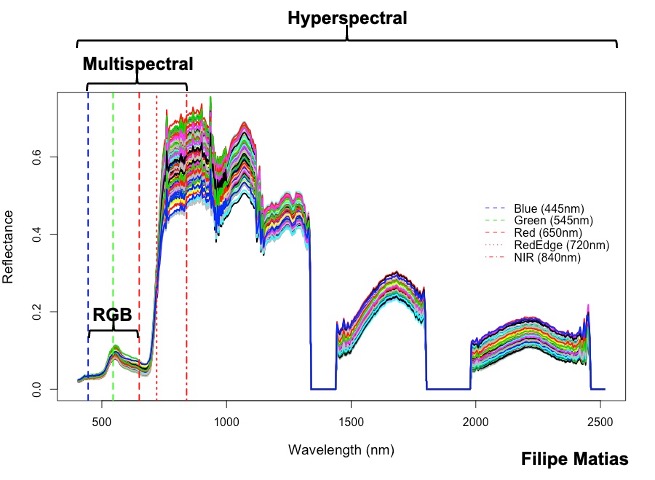
[Menu](#menu)
---------------------------------------------
#### 14. Building shapefile with polygons (field blocks, pest damage, soil differences, etc)
> If your field area does not have a pattern to draw plots using the function **fieldShape** you can draw different polygons using the function **fieldPolygon**. To make the fieldshape file the number of polygons must be informed. For instance, for each polygon, you should select at least four points at the polygon boundaries area. This function is recommended to make shapefile to extract data from specific field blocks, pest damage area, soil differences, etc. If *extent=TRUE* the whole image area will be the shapefile (used to analyze multiple images for example to evaluate seeds, ears, leaves, diseases, etc.). Function to use: **fieldPolygon**. The following example uses an image available to download here: [EX_polygonShape.tif](https://drive.google.com/open?id=1b42EAi3A3X54z00JWjFvsmUAm3yQClPk).
```r
# Uploading file (EX_polygonShape.tif)
EX.polygon<-rast("EX_polygonShape.tif")
plotRGB(EX.polygon, r = 1, g = 2, b = 3)
# Removing soil
EX.polygon.RemSoil<- fieldMask(mosaic = EX.polygon)
# Data frame with polygons information
polygonData<-data.frame(ID=c("Polygon1","Polygon2","Polygon3"),
FlowerColor=c("white","white","white"),
FlowerPercent=c(20,40,50),
LeafColor=c("dark","light","dark"))
polygonData
# Building plot shapefile with 3 polygons (select 4 points around the polygon area)
EX.polygon.Shape<-fieldPolygon(mosaic = EX.polygon.RemSoil,
nPolygon = 3,nPoint = 4,ID = "ID",
polygonData = polygonData,cropPolygon = T,
polygonID = c("Polygon1","Polygon2","Polygon3"))
plotRGB(EX.polygon.Shape$cropField)
# Building indice (NGRDI and BGI)
EX.polygon.Indices<- fieldIndex(mosaic = EX.polygon.RemSoil$newMosaic, Red = 1, Green = 2, Blue = 3,
index = c("NGRDI","BGI"))
# Extracting data (NGRDI and BGI)
EX.polygon.Info<- fieldInfo_extra(mosaic = EX.polygon.Indices[[c("NGRDI","BGI")]],
fieldShape = EX.polygon.Shape)
EX.polygon.Info
# Making graphics (BGI)
fieldPlot(fieldShape=EX.polygon.Info,
fieldAttribute="BGI",
mosaic=EX.polygon, color=c("red","blue"), alpha = 0.5)
```
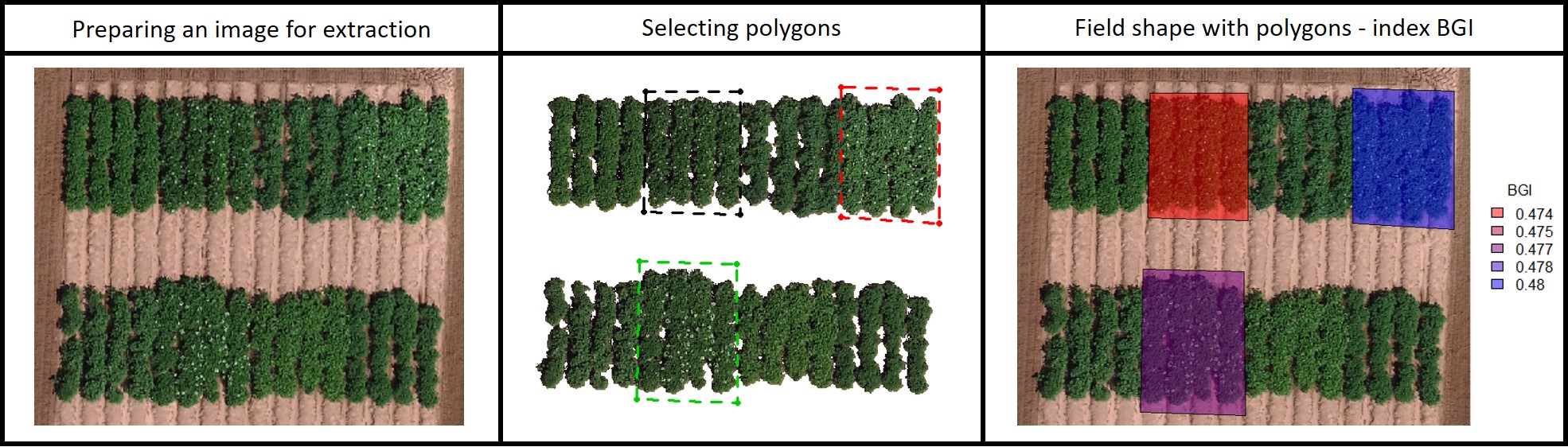
[Menu](#menu)
---------------------------------------------
#### 15. Making plots
> Graphic visualization of trait values for each plot using the **fieldShape file** and the **Mosaic** of your preference. Function to use: **`fieldPlot`**.
```r
### Interpolating colors: c("white","black")
fieldPlot(fieldShape=EX1.Info$fieldShape,fieldAttribute="Yield", mosaic=EX1.Indices, color=c("white","black"), alpha = 0.5)
### Interpolating colors: c("red","blue")
fieldPlot(fieldShape=EX1.Info$fieldShape,fieldAttribute="myIndex", mosaic=EX1.Indices, color=c("red","blue"), alpha = 0.5)
```
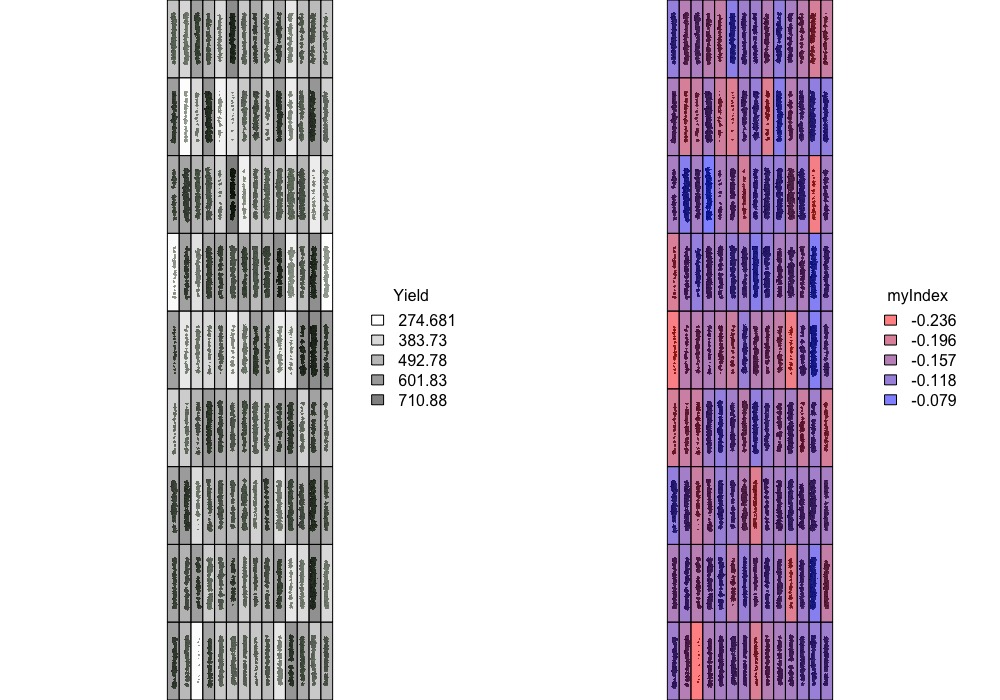
[Menu](#menu)
---------------------------------------------
#### 16. Saving output files
```r
### Images (single and multi layers)
writeRaster(EX1.Indices, filename="EX1.Indices.tif", overwrite=TRUE)
# EX1.Indices.2 <- rast("EX1.Indices.tif") # Reading the saved image.
### FieldShape file
st_write(EX1.Info$fieldShape, ".", "EX1.fieldShape", driver="ESRI Shapefile")
# EX1.fieldShape.2 <- st_read("EX1.fieldShape.shp") # Reading the saved shapefile option 01.
### CSV file (table)
write.csv(EX1.Info,file = "EX1.Info.csv",col.names = T,row.names = F)
# Data.EX1.Info<-read.csv("EX1.Info.csv",header = T,check.names = F) # Reading the saved data table.
```
[Menu](#menu)
---------------------------------------------
#### Orthomosaic using the open source software **[OpenDroneMap](https://www.opendronemap.org)**
> Image stitching from remote sensing phenotyping platforms (sensors attached to aerial or ground vehicles) in one orthophoto.
1) Follow the [OpenDroneMap’s documentation](https://docs.opendronemap.org/index.html) according to your operating system (Windows, macOS or Linux) to install [**WebODM**](https://www.opendronemap.org/webodm/).
2) Start [WebODM](https://www.opendronemap.org/webodm/) and **+Add Project** to upload your pictures (.tif or .jpg). Attached is one example of RGB images from experimental trials of [UW-Madison Potato Breeding and Genetics Laboratory](https://potatobreeding.cals.wisc.edu) during the flowering time at [Hancock Agricultural Research Station](https://hancock.ars.wisc.edu). Flight altitude was 60 m above ground, flight speed was 24 km/h, and image overlap was 75%. Donwload pictures [here](https://drive.google.com/open?id=1t0kjcBy6QzmIz_fVs6vsgZXi9Afqe09b).
3) After the running process '*completed*', download the **odm_orthophoto.tif** and **dsm.tif** to upload in R. Then follow the pipeline of *FIELDimageR*. [Donwload the final *odm_orthophoto.tif](https://drive.google.com/open?id=1XCDvbFdzHDmRA1dJSMp_veX_xC_ThiqC)*.
```r
# Uploading file (odm_orthophoto.tif):
EX.ODM<-rast("odm_orthophoto.tif")
plotRGB(EX.ODM, r = 1, g = 2, b = 3)
# Cropping the image to select only one trial (Selecting the same trial as EX.2 from step.13):
EX.ODM.Crop <- fieldCrop(mosaic = EX.ODM)
# Rotating the image using the same theta from step 3:
EX.ODM.Rotated<-fieldRotate(EX.ODM.Crop,theta = 2.3)
# Removing soil
EX.ODM.RemSoil<- fieldMask(mosaic = EX.ODM.Rotated)
# Building indices
EX.ODM.Indices <- fieldIndex(EX.ODM.RemSoil$newMosaic,Red=1,Green=2,Blue=3,
index = c("NGRDI","BGI"), myIndex = c("(Red-Blue)/Green"))
# Extracting data using the same fieldShape file from step 5:
EX.ODM.Info<- fieldInfo_extra(mosaic = EX.ODM.Indices$myIndex,fieldShape = EX1.Shape)
EX.ODM.Info$myIndex
```

[Menu](#menu)
---------------------------------------------
#### Parallel and loop to evaluate multiple images
> The following code can be used to evaluate multiple images (e.g. root area, leave indices, damaged area, ears, seed, structures, etc.). The example below is evaluating disease damage in images of leaves (affected area and indices). Download the example [here](https://drive.google.com/open?id=1zgOZFd7KuTu4sERcG1wFoAWLCahIJIlu).
```r
# Images names (folder directory: "./images/")
pics<-list.files("./images/")
# Vegetation indices
index<- c("BGI","VARI","SCI")
############
### Loop ###
############
system.time({ # system.time: used to compare the processing time using loop and parallel
EX.Table.Loop<-NULL
for(i in 1:length(pics)){
EX.L1<-rast(paste("./images/",pics[i],sep = ""))
plotRGB(EX.L1)
EX.L.Shape<-fieldPolygon(mosaic=EX.L1, extent=T, plot=F) # extent=T (The whole image area will be the shapefile)
EX.L2<-fieldMask(mosaic=EX.L1, index="BGI", cropValue=0.8, cropAbove=T, plot=F) # Select one index to identify leaves and remove the background
EX.L3<-fieldMask(mosaic=EX.L2$newMosaic, index="VARI", cropValue=0.1, cropAbove=T, plot=F) # Select one index to identify demaged area in the leaves
EX.L4<-fieldIndex(mosaic=EX.L2$newMosaic, index=index, plot=F) # Indices
EX.L5<-rast(EX.L3$mask, EX.L4[[index]]) # Making a new rast raster with new layers (demage area and indices)
EX.L.Info<- fieldInfo_extra(mosaic=EX.L5, fieldShape=EX.L.Shape, projection=F) # projection=F (Ignore projection. Normally used only with remote sensing images)
plot(EX.L5,col = grey(1:100/100))
EX.Table.Loop<-rbind(EX.Table.Loop, EX.L.Info$plotValue) # Combine information from all images in one table
}})
rownames(EX.Table.Loop)<-pics
EX.Table.Loop
################
### Parallel ###
################
# Required packages
library(parallel)
library(foreach)
library(doParallel)
# Number of cores
n.core<-detectCores()-1
# Starting parallel
cl <- makeCluster(n.core, output = "")
registerDoParallel(cl)
system.time({
EX.Table.Parallel <- foreach(i = 1:length(pics), .packages = c("raster","FIELDimageR"),
.combine = rbind) %dopar% {
EX.L1<-rast(paste("./images/",pics[i],sep = ""))
EX.L.Shape<-fieldPolygon(mosaic=EX.L1, extent=T, plot=F) # extent=T (The whole image area will be the shapefile)
EX.L2<-fieldMask(mosaic=EX.L1, index="BGI", cropValue=0.8, cropAbove=T, plot=F) # Select one index to identify leaves and remove the background
EX.L3<-fieldMask(mosaic=EX.L2$newMosaic, index="VARI", cropValue=0.1, cropAbove=T, plot=F) # Select one index to identify demaged area in the leaves
EX.L4<-fieldIndex(mosaic=EX.L2$newMosaic, index=index, plot=F) # Indices
EX.L5<-rast(EX.L3$mask, EX.L4[[index]]) # Making a new rast raster with new layers (demage area and indices)
EX.L.Info<- fieldInfo_extra(mosaic=EX.L5, fieldShape=EX.L.Shape, projection=F) # projection=F (Ignore projection. Normally used only with remote sensing images)
EX.L.Info$plotValue # Combine information from all images in one table
}})
stopCluster(cl)
rownames(EX.Table.Parallel)<-pics
EX.Table.Parallel
```

[Menu](#menu)
---------------------------------------------
#### Quick tips (image analyze in R)
1) [Changing memory limits in R](https://stat.ethz.ch/R-manual/R-devel/library/base/html/Memory-limits.html)
```
# The rasterOptions() allows you to customize your R session (raster package):
rasterOptions()
rasterOptions(chunksize = 1e+09)
rasterOptions(maxmemory = 1e+09)
```
2) [Reducing resolution for fast analysis](https://www.rdocumentation.org/packages/raster/versions/3.0-12/topics/aggregate)
```
new_mosaic<-aggregate(previous_mosaic, fact=4)
```
3) Removing previous files after used (more space in R memory): If you do not need one mosaic you should remove it to save memory to the next steps (e.g. the first input mosaic)
```
rm(previous_mosaic)
```
4) Using ShapeFile from other software (e.g. [QGIS](https://www.qgis.org/en/site/))
```
ShapeFile <- rgdal::readOGR("Other_Software_ShapeFile.shp") # Option 01
ShapeFile <- terra::vect("Other_Software_ShapeFile.shp") # Option 02
```
5) Combining ShapeFiles: sometimes it is better to split the mosaic into smaller areas to better draw the shapefile. For instance, the user can combine the split shapefiles to one for the next step as extractions
```
ShapeFile <- rbind(ShapeFile1, ShapeFile2, ShapeFile3, ...)
```
6) External window to amplify the RStudio plotting area (It helps to visualize and click when using functions: **fieldCrop**, **fieldRotate**, **fieldShape**, and **fieldPolygon**). The default graphics device is normally "RStudioGD". To change use "windows" on Windows, "quartz" on MacOS, and "X11" on Linux.
```
# Example in macOS (type the following code before running FIELDimageR functions):
options(device = "quartz")
```
[Menu](#menu)
---------------------------------------------
### YouTube Tutorial
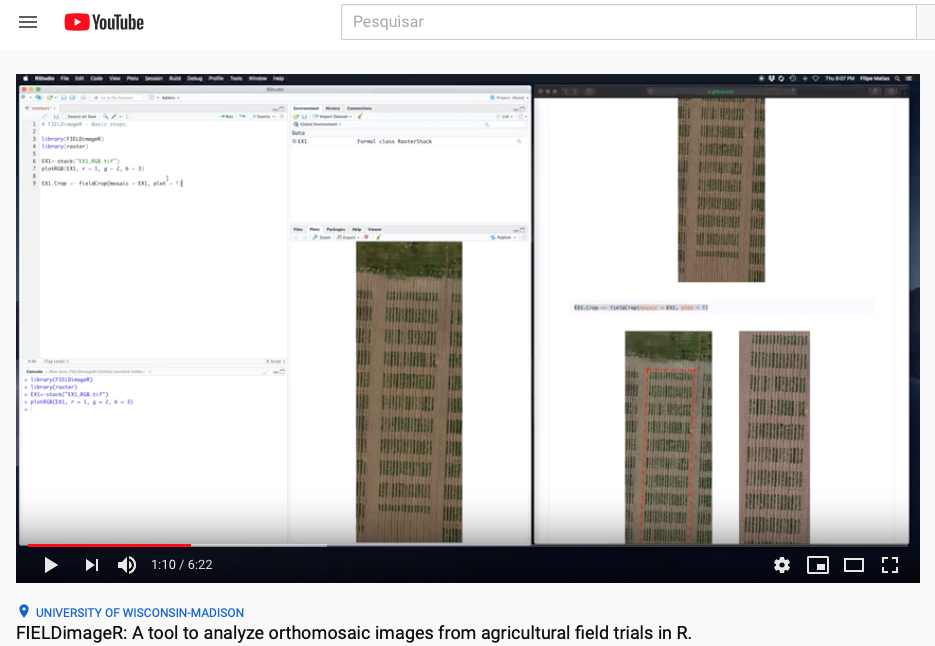
> FIELDimageR: A tool to analyze orthomosaic images from agricultural field trials in R (Basic Pipeline)
> FIELDimageR: Counting the number of plants (fieldCount)
> FIELDimageR: Calculating vegetation indices (NDVI and NDRE)
> FIELDimageR: Estimate plant height using the canopy height model (CHM)
### FIELDimageR courses
* **Embrapa Beef-Cattle**: https://fieldimager.cnpgc.embrapa.br/course.html
* **Embrapa Maize & Sorghum**: http://fieldimager.cnpms.embrapa.br/course.html
* **Universidade Federal de Viçosa (GenMelhor)**: https://genmelhor-fieldimager.000webhostapp.com/FIELDimageR_Course%20(2).html
* **PhenomeForce (Fridays Hands-On: A Workshop Series in Data+Plant Sciences)**: https://phenome-force.github.io/FIELDimageR-workshop/
* **The Plant Phenome Journal**: https://www.youtube.com/watch?v=DOD0ZX_J8tk
### Google Groups Forum
> This discussion group provides an online source of information about the FIELDimageR package. Report a bug and ask a question at:
* https://groups.google.com/forum/#!forum/fieldimager
* https://community.opendronemap.org/t/about-the-fieldimager-category/4130
### Developers
> **Help improve FIELDimageR pipeline**. The easiest way to modify the package is by cloning the repository and making changes using [R projects](https://support.rstudio.com/hc/en-us/articles/200526207-Using-Projects).
> If you have questions, join the forum group at https://groups.google.com/forum/#!forum/fieldimager
>Try to keep commits clean and simple
>Submit a **_pull request_** with detailed changes and test results.
**Let's work together and help more people (students, professors, farmers, etc) to have access to this knowledge. Thus, anyone anywhere can learn how to apply remote sensing in agriculture.**
### Licenses
> The R/FIELDimageR package as a whole is distributed under [GPL-2 (GNU General Public License version 2)](https://www.gnu.org/licenses/gpl-2.0.en.html).
### Citation
> *Matias FI, Caraza-Harter MV, Endelman JB.* FIELDimageR: An R package to analyze orthomosaic images from agricultural field trials. **The Plant Phenome J.** 2020; [https://doi.org/10.1002/ppj2.20005](https://doi.org/10.1002/ppj2.20005)
> * Pawar P & Matias FI.* FIELDimageR.Extra. *Pawar P. & Matias FI.* FIELDimageR.Extra: Advancing user experience and computational efficiency for analysis of orthomosaic from agricultural field trials. **The Plant Phenome J.** 2023; [https://doi.org/10.1002/ppj2.20083](https://doi.org/10.1002/ppj2.20083)
### Author
> * [Filipe Inacio Matias](https://github.com/filipematias23)
### Acknowledgments
> * [University of Wisconsin - Madison](https://horticulture.wisc.edu)
> * [UW Potato Breeding and Genetics Laboratory](https://potatobreeding.cals.wisc.edu)
> * [Dr Jeffrey Endelman, PhD Student Maria Caraza-Harter, and MS Student
Lin Song](https://potatobreeding.cals.wisc.edu/people/)
> * [OpenDroneMap](https://www.opendronemap.org/)
[Menu](#menu)