https://github.com/brunobrr/bdc
Check out the vignettes with detailed documentation on each module of the bdc package
https://github.com/brunobrr/bdc
bdc biodiversity-data workflow
Last synced: 4 months ago
JSON representation
Check out the vignettes with detailed documentation on each module of the bdc package
- Host: GitHub
- URL: https://github.com/brunobrr/bdc
- Owner: brunobrr
- License: gpl-3.0
- Created: 2020-09-29T18:48:10.000Z (over 5 years ago)
- Default Branch: master
- Last Pushed: 2025-05-16T11:27:07.000Z (9 months ago)
- Last Synced: 2025-10-07T22:01:58.815Z (5 months ago)
- Topics: bdc, biodiversity-data, workflow
- Language: R
- Homepage: https://brunobrr.github.io/bdc
- Size: 179 MB
- Stars: 24
- Watchers: 3
- Forks: 9
- Open Issues: 7
-
Metadata Files:
- Readme: README.Rmd
- Changelog: NEWS.md
- License: LICENSE.md
Awesome Lists containing this project
- open-sustainable-technology - bdc - A toolkit for standardizing, integrating, and cleaning biodiversity data. (Biosphere / Biodiversity Data Cleaning and Standardization)
README
---
output: github_document
editor_options:
markdown:
wrap: 80
---
```{r, include = FALSE}
knitr::opts_chunk$set(
collapse = TRUE,
comment = "#>",
fig.path = "man/figures/README-",
out.width = "100%"
)
```
## **A toolkit for standardizing, integrating, and cleaning biodiversity data**
[](https://CRAN.R-project.org/package=bdc)
[](https://cranlogs.r-pkg.org:443/badges/grand-total/bdc)
[](https://github.com/brunobrr/bdc/actions/workflows/R-CMD-check.yaml)
[](https://app.codecov.io/gh/brunobrr/bdc)
[](https://doi.org/10.5281/zenodo.6450390)
[-lightgrey.svg?style=flat)](http://www.gnu.org/licenses/gpl-3.0.html)
#### **Overview**
Handle biodiversity data from several different sources is not an easy task.
Here, we present the **B**iodiversity **D**ata **C**leaning (*bdc*), an R
package to address quality issues and improve the fitness-for-use of biodiversity
datasets. *bdc* contains functions to harmonize and integrate data from
different sources following common standards and protocols, and implements
various tests and tools to flag, document, clean, and correct taxonomic,
spatial, and temporal data.
Compared to other available R packages, the main strengths of the *bdc* package
are that it brings together available tools – and a series of new ones – to
assess the quality of different dimensions of biodiversity data into a single
and flexible toolkit. The functions can be applied to a multitude of taxonomic
groups, datasets (including regional or local repositories), countries, or
worldwide.
#### **Structure of *bdc***
The *bdc* toolkit is organized in thematic modules related to different
biodiversity dimensions.
--------------------------------------------------------------------------------
> :warning: The modules illustrated, and **functions** within, **were linked to
> form** a proposed reproducible **workflow** (see
> [**vignettes**](https://brunobrr.github.io/bdc/)). However, all functions
> **can also be executed independently**.
--------------------------------------------------------------------------------
#### 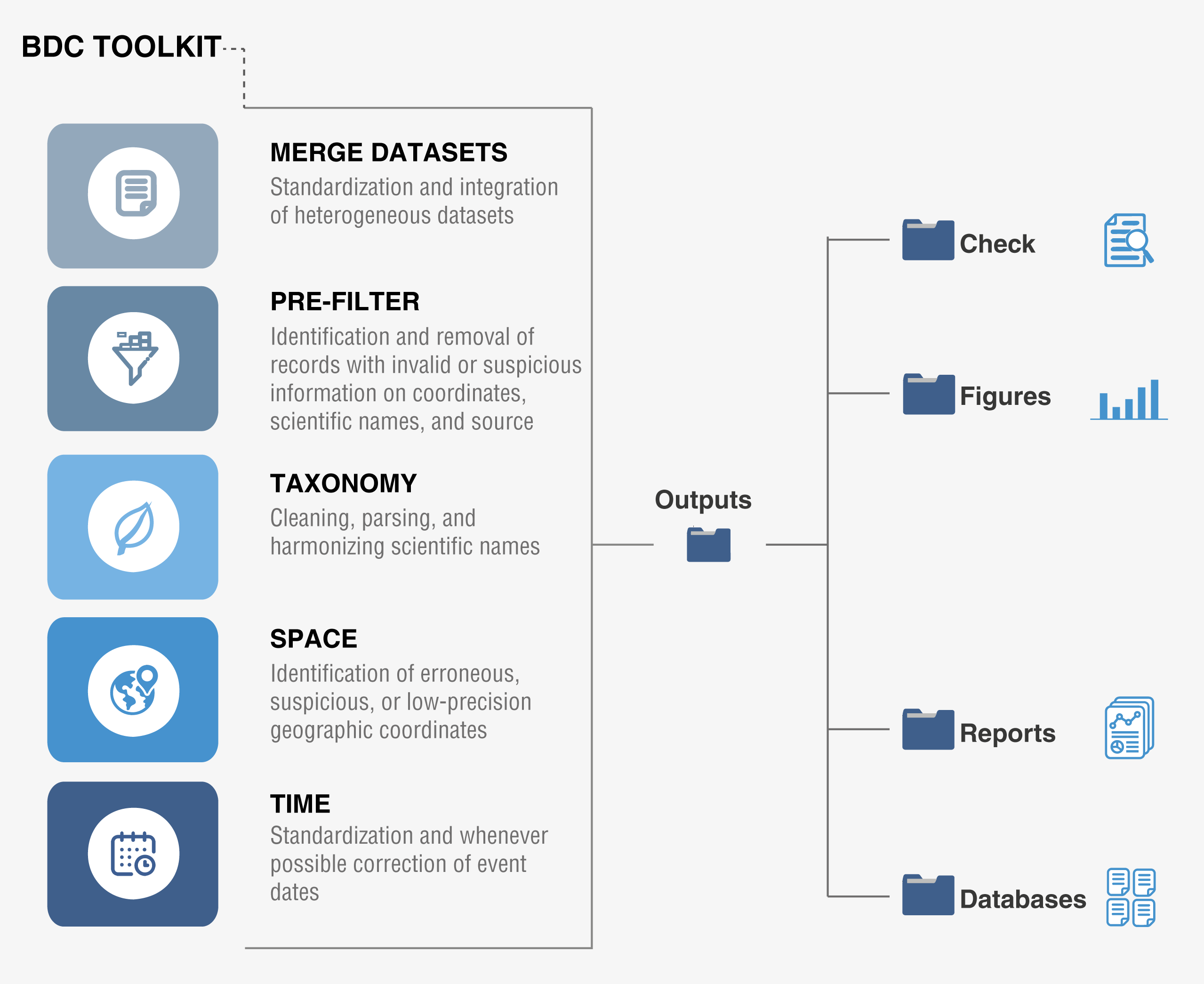
#### 1. [**Merge databases**](https://brunobrr.github.io/bdc/articles/integrate_datasets.html)
Standardization and integration of different datasets into a standard database.
- `bdc_standardize_datasets()` Standardization and integration of different
datasets into a new dataset with column names following Darwin Core
terminology
#### 2. [**Pre-filter**](https://brunobrr.github.io/bdc/articles/prefilter.html)
Flagging and removal of invalid or non-interpretable information, followed by
data amendments (e.g., correct transposed coordinates and standardize country
names).
- `bdc_scientificName_empty()` Identification of records lacking names or with
names not interpretable
- `bdc_coordinates_empty()` Identification of records lacking information on
latitude or longitude
- `bdc_coordinates_outOfRange()` Identification of records with out-of-range
coordinates (latitude \> 90 or -90; longitude \>180 or -180)
- `bdc_basisOfRecords_notStandard()` Identification of records from doubtful
sources (e.g., fossil or machine observation) impossible to interpret and
not compatible with Darwin Core recommended vocabulary
- `bdc_country_from_coordinates()` Derive country name from valid geographic
coordinates
- `bdc_country_standardized()` Standardization of country names and retrieve
country code
- `bdc_coordinates_transposed()` Identification of records with potentially
transposed latitude and longitude
- `bdc_coordinates_country_inconsistent()` Identification of coordinates in
other countries or far from a specified distance from the coast of a
reference country (i.e., in the ocean)
- `bdc_coordinates_from_locality()` Identification of records lacking
coordinates but with a detailed description of the locality associate with
records from which coordinates can be derived
#### 3. [**Taxonomy**](https://brunobrr.github.io/bdc/articles/taxonomy.html)
Cleaning, parsing, and harmonization of scientific names against multiple
taxonomic references.
- `bdc_clean_names()` Name-checking routines to clean and split a taxonomic
name into its binomial and authority components
- `bdc_query_names_taxadb()` Harmonization of scientific names by correcting
spelling errors and converting nomenclatural synonyms to currently accepted
names.
- `bdc_filter_out_names()` Function used to filter out records according to
their taxonomic status present in the column "notes". For example, to filter
only valid accepted names categorized as "accepted"
#### 4. [**Space**](https://brunobrr.github.io/bdc/articles/space.html)
Flagging of erroneous, suspicious, and low-precision geographic coordinates.
- `bdc_coordinates_precision()` Identification of records with a coordinate
precision below a specified number of decimal places
- `clean_coordinates()` (From *CoordinateCleaner* package and part of the
data-cleaning workflow). Identification of potentially problematic
geographic coordinates based on geographic gazetteers and metadata. Include
tests for flagging records: around country capitals or country or province
centroids, duplicated, with equal coordinates, around biodiversity
institutions, within urban areas, plain zeros in the coordinates, and
suspect geographic outliers
#### 5. [**Time**](https://brunobrr.github.io/bdc/articles/time.html)
Flagging and, whenever possible, correction of inconsistent collection date.
- `bdc_eventDate_empty()` Identification of records lacking information on
event date (i.e., when a record was collected or observed)
- `bdc_year_outOfRange()` Identification of records with illegitimate or
potentially imprecise collecting year. The year provided can be out-of-range
(e.g., in the future) or collected before a specified year supplied by the
user (e.g., 1900)
- `bdc_year_from_eventDate()` This function extracts four-digit year from
unambiguously interpretable collecting dates
#### [**Other functions**](https://brunobrr.github.io/bdc/reference/index.html)
Aim to facilitate the **documentation, visualization, and interpretation** of
results of data quality tests the package contains functions for documenting the
results of the data-cleaning tests, including functions for saving i) records
needing further inspection, ii) figures, and iii) data-quality reports.
- `bdc_create_report()` Creation of data-quality reports documenting the
results of data-quality tests and the taxonomic harmonization process
- `bdc_create_figures()` Creation of figures (i.e., bar plots and maps)
reporting the results of data-quality tests
- `bdc_filter_out_flags()` Removal of columns containing the results of data
quality tests (i.e., column starting with ".") or other columns specified
- `bdc_quickmap()` Creation of a map of points using ggplot2. Helpful in
inspecting the results of data-cleaning tests
- `bdc_summary_col()` This function creates or updates the column summarizing
the results of data quality tests (i.e., the column ".summary")
#### **Installation**
```{r eval=FALSE}
install.packages("bdc")
library(bdc)
```
or the development version from [GitHub](https://github.com/brunobrr/bdc) using:
```{r, message=FALSE, warning=FALSE,echo=TRUE,eval=FALSE}
install.packages("remotes")
remotes::install_github("brunobrr/bdc")
```
Load the package with:
```{r, message=FALSE, warning=FALSE,echo=TRUE,eval=TRUE}
library(bdc)
```
#### **Package website**
See *bdc* package website () for detailed
explanation on each module.
#### **Getting help**
> If you encounter a clear bug, please file an issue
> [**here**](https://github.com/brunobrr/bdc/issues). For questions or
> suggestion, please send us a email (ribeiro.brr\@gmail.com).
#### **Citation**
Ribeiro, BR; Velazco, SJE; Guidoni-Martins, K; Tessarolo, G; Jardim, Lucas;
Bachman, SP; Loyola, R (2022). bdc: A toolkit for standardizing, integrating,
and cleaning biodiversity data. Methods in Ecology and Evolution.
[doi.org/10.1111/2041-210X.13868](https://doi.org/10.1111/2041-210X.13868)
