https://github.com/ropensci/mregions2
Access the Marine Regions Gazetteer and the Marine Regions Data Products in R. Maintained by @salvafern.
https://github.com/ropensci/mregions2
Last synced: about 1 month ago
JSON representation
Access the Marine Regions Gazetteer and the Marine Regions Data Products in R. Maintained by @salvafern.
- Host: GitHub
- URL: https://github.com/ropensci/mregions2
- Owner: ropensci
- License: other
- Created: 2022-04-25T10:27:29.000Z (over 3 years ago)
- Default Branch: main
- Last Pushed: 2025-03-24T14:56:49.000Z (8 months ago)
- Last Synced: 2025-10-17T15:27:43.860Z (about 1 month ago)
- Language: R
- Homepage: https://docs.ropensci.org/mregions2/
- Size: 7.02 MB
- Stars: 9
- Watchers: 4
- Forks: 4
- Open Issues: 0
-
Metadata Files:
- Readme: README.Rmd
- Changelog: NEWS.md
- Contributing: .github/CONTRIBUTING.md
- License: LICENSE
- Codemeta: codemeta.json
Awesome Lists containing this project
- open-sustainable-technology - mregions2 - Offers a streamlined interface to access data from Marine Regions in R for researchers, marine scientists, and geospatial analysts seeking marine geographical information. (Hydrosphere / Ocean and Hydrology Data Access)
- jimsghstars - ropensci/mregions2 - Access the Marine Regions Gazetteer and the Marine Regions Data Products in R. Maintained by @salvafern. (R)
README
---
output: github_document
df_print: tibble
---
```{r, include = FALSE}
knitr::opts_chunk$set(
collapse = TRUE,
comment = "#>",
fig.path = "man/figures/README-",
out.width = "100%",
cache = TRUE
)
```
# mregions2
[](https://lifewatch.be) [](https://github.com/ropensci/software-review/issues/590) [](https://cran.r-project.org/package=mregions2) [](https://github.com/ropensci/mregions2/actions/workflows/R-CMD-check.yaml) [](https://app.codecov.io/gh/ropensci/mregions2?branch=main)

`mregions2` offers a streamlined interface to access data from [Marine Regions](https://marineregions.org) in R for researchers, marine scientists, and geospatial analysts seeking marine geographical information
Marine Regions offers two key resources: the [Marine Regions Gazetteer](https://marineregions.org/gazetteer.php), a list of standardized marine place names with unique identifiers, and the [Marine Regions Data Products](https://marineregions.org/sources.php), including popular features like the world maritime boundaries.
You can find detailed information in the articles online:
- [Introduction to mregions2](https://docs.ropensci.org/mregions2/articles/mregions2.html)
- [Why mregions and mregions2?](https://docs.ropensci.org/mregions2/articles/why_mregions2.html)
- [Marine Regions Data Products Ontology](https://docs.ropensci.org/mregions2/articles/mrp_ontology.html)
- [mregions2 as RDF](https://docs.ropensci.org/mregions2/articles/mregions2-rdf.html)
## Installation
You can install the latest CRAN version with:
``` r
install.packages("mregions2")
```
Or install the development version of mregions2 from GitHub with:
``` r
# install.packages("devtools")
devtools::install_github("ropensci/mregions2", build_vignettes = TRUE)
```
Load the library with:
```{r, results='hide', warning=FALSE, message=FALSE}
library(mregions2)
```
The function `mrp_view()` requires two extra packages that are not listed as Imports, hence they are not installed along with `mregions2` but you must install yourself:
``` r
install.packages("leaflet")
install.packages("leaflet.extras2")
```
Some of the examples below use the pipe operator `%>%`. Install and load `magrittr`:
```{r}
# install.packages("magrittr")
library(magrittr)
```
## Query the Marine Regions Gazetteer
The [Marine Regions Gazetteer](https://marineregions.org/gazetteer.php) is a standard list of marine georeferenced place names.
> Gazetteer: a dictionary of geographical names.
You can search the Gazetteer in many ways:
Search by free text:
```{r}
gaz_search("Belgian Part of the North Sea")
```
Search by unique identifier. See `?MRGID`:
```{r}
gaz_search(3293)
```
Search by location:
```{r}
gaz_search(x = 2.927, y = 51.21551)
```
Search by place type:
```{r}
gaz_search_by_type("EEZ")
```
Search by authortative source:
```{r}
gaz_search_by_source("Flanders Marine Institute (VLIZ)")
```
The list of place types and sources are available with `gaz_types()` and `gaz_sources()` respectively.
You can add the geometry of the Gazetteer entries with `gaz_geometry()`:
```{r gaz2}
# Get a record and turn into a sf object with geometry
gaz_search(3293) %>% gaz_geometry()
# Or get only the geometry
gaz_geometry(3293, format = "sfc")
```
The entries of the Marine Regions Gazetteer are organized hierarchically. You can browse this hierarchy up and down with `gaz_relations()`
```{r gaz3}
# Get all relations
gaz_search(3293) %>% gaz_relations()
# Or get the relations directly
gaz_relations(3293)
```
## Marine Regions Data Products
In addition to the Marine Regions Gazetteer, the Marine Regions Team creates and hosts geographical Data Products, being the most popular one the [Marine Regions Maritime Boundaries](https://marineregions.org/eez.php).
An overview of all available products can be consulted with `mrp_list`
```{r prod_list}
mrp_list
```
Their attributes are explained in the [Marine Regions Data Products Ontology article](https://docs.ropensci.org/mregions2/articles/mrp_ontology.html), or simply run `mrp_ontology`
```{r prod_ontology}
mrp_ontology
```
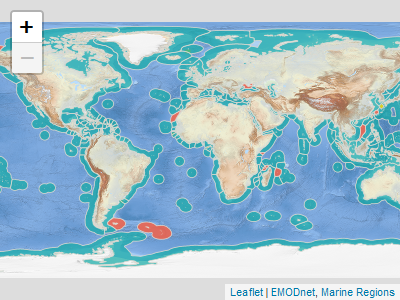
You can visualize the Marine Regions Data Products with `mrp_view()`. It opens an interactive `leaflet::leaflet` viewer.
```{r prod1, eval=FALSE}
mrp_view("eez")
```
Or you can download and read the data products into R with `mrp_get()`
```{r prod2, eval = FALSE}
mrp_get("eez")
```
You can specify the download path in the `path` argument:
```{r prod3, eval = FALSE}
mrp_get("eez", path = "path/to/data")
```
Get to know more in the [Get Started vignette](https://docs.ropensci.org/mregions2/articles/mregions2.html).
```{r, include=FALSE, echo=FALSE}
# Copy and paste output - do not include to not add more deps
get_md <- function(pkgs){
get_titles <- function(x) packageDescription(x, fields = "Title")
titles <- purrr::map_chr(pkgs, get_titles)
cat(glue::glue("* [{pkgs}](https://docs.ropensci.org/{pkgs}): _{titles}_. \n"), sep = "\n")
}
```
## Related packages
### Marine biodiversity data
The main purpose of Marine Regions is to serve as the geographical backbone for the World Register of Marine Species (WoRMS), an authoritative classification and catalogue of marine names. Here is a list of R packages to access Marine taxonomical and biogeographic data that can be combined with data from `mregions2`:
```{r, include=FALSE, echo=FALSE}
get_md(c("worrms", "eurobis", "robis", "rgbif"))
# Change links manually
```
* [worrms](https://docs.ropensci.org/worrms/): _World Register of Marine Species (WoRMS) Client_.
* [eurobis](https://lifewatch.github.io/eurobis/): _Download data from EurOBIS using the LifeWatch/EMODnet-Biology Web Feature Services_.
* [robis](https://cran.r-project.org/package=robis): _Ocean Biodiversity Information System (OBIS) Client_.
* [rgbif](https://docs.ropensci.org/rgbif/): _Interface to the Global Biodiversity Information Facility API_.
### More geographical data
There are other R packages that allow to access other gazetteers and boundaries data. Here is a non-exhaustive list:
```{r, include=FALSE, echo=FALSE}
get_md(c("geonames", "osmdata", "osmextract", "rnaturalearth"))
```
* [geonames](https://docs.ropensci.org/geonames/): _Interface to the "Geonames" Spatial Query Web Service_.
* [osmdata](https://docs.ropensci.org/osmdata/): _Import 'OpenStreetMap' Data as Simple Features or Spatial
Objects_.
* [osmextract](https://docs.ropensci.org/osmextract/): _Download and Import Open Street Map Data Extracts_.
* [rnaturalearth](https://docs.ropensci.org/rnaturalearth/): _World Map Data from Natural Earth_.
## Citation
```{r citation, cache=FALSE, collapse=FALSE, comment="", results='asis'}
citation("mregions2")
```
## License
MIT. See `LICENSE.md`
This software is developed for scientific, educational and research purposes. It is not meant to be used for legal, economical (in the sense of exploration of natural resources) or navigational purposes. See the [Marine Regions disclaimer](https://marineregions.org/disclaimer.php) for more information.
## Code of Conduct
Please note that this package is released with a [Contributor Code of Conduct](https://ropensci.org/code-of-conduct/). By contributing to this project, you agree to abide by its terms.