https://github.com/viswanath-lab/RadQy
MRQy is a quality assurance and checking tool for quantitative assessment of magnetic resonance imaging (MRI) data.
https://github.com/viswanath-lab/RadQy
imaging machine-learning medical-image-processing medical-imaging medical-physics mri python quality-assurance quality-control quality-metrics
Last synced: 5 months ago
JSON representation
MRQy is a quality assurance and checking tool for quantitative assessment of magnetic resonance imaging (MRI) data.
- Host: GitHub
- URL: https://github.com/viswanath-lab/RadQy
- Owner: ccipd
- License: bsd-3-clause-clear
- Created: 2019-10-08T12:49:45.000Z (about 6 years ago)
- Default Branch: master
- Last Pushed: 2024-03-16T14:03:53.000Z (over 1 year ago)
- Last Synced: 2024-05-08T05:41:45.844Z (over 1 year ago)
- Topics: imaging, machine-learning, medical-image-processing, medical-imaging, medical-physics, mri, python, quality-assurance, quality-control, quality-metrics
- Language: JavaScript
- Homepage:
- Size: 229 MB
- Stars: 88
- Watchers: 5
- Forks: 27
- Open Issues: 7
-
Metadata Files:
- Readme: README.md
- Contributing: ContributingGuidelines.md
- License: LICENSE.txt
Awesome Lists containing this project
- awesoMRI-QC - MRQy
README

## Front-end View

## Backend View
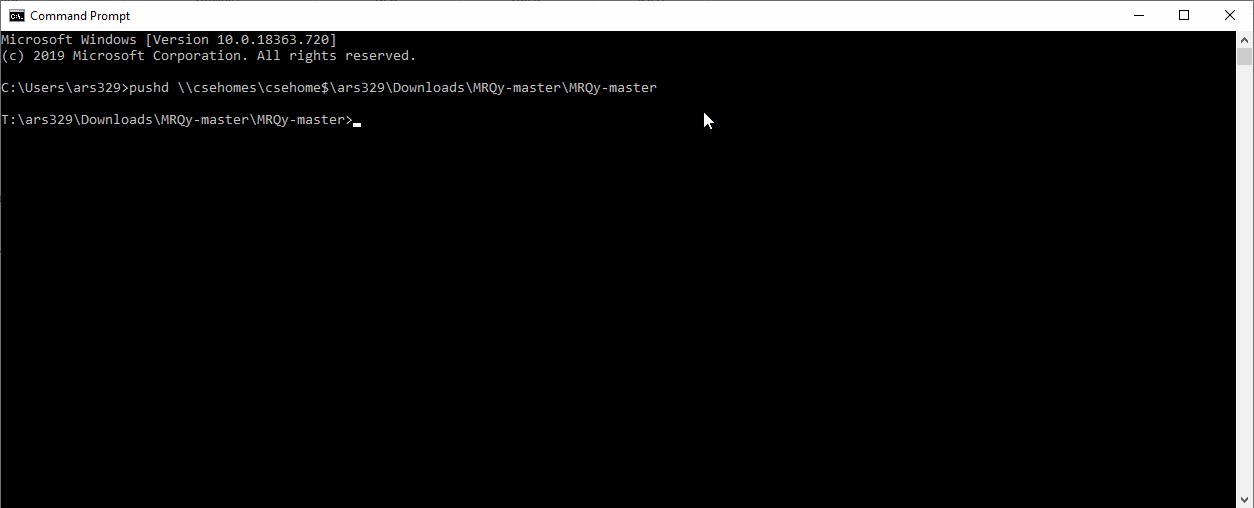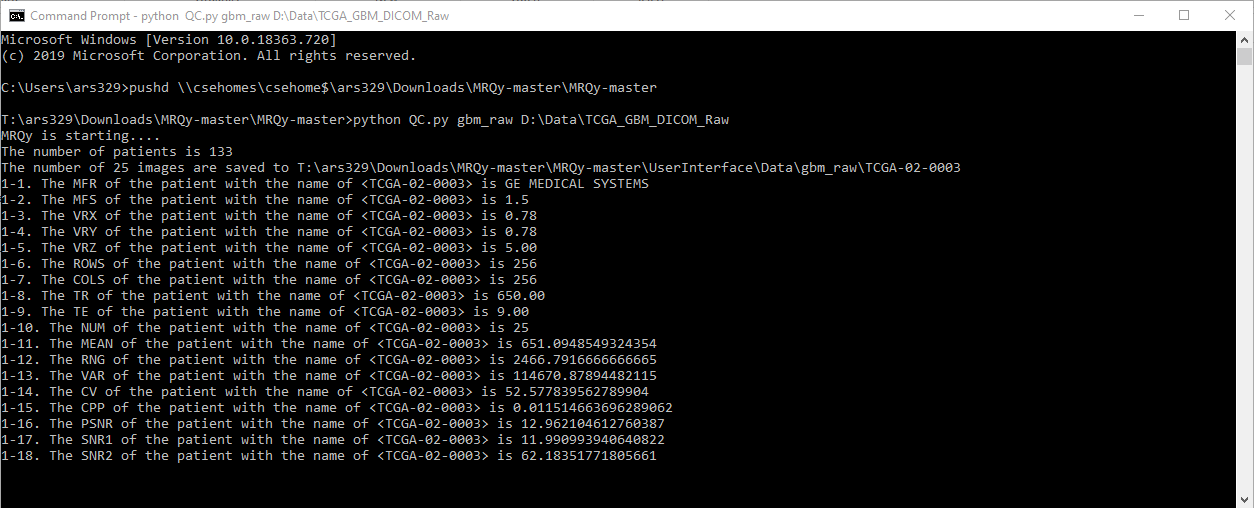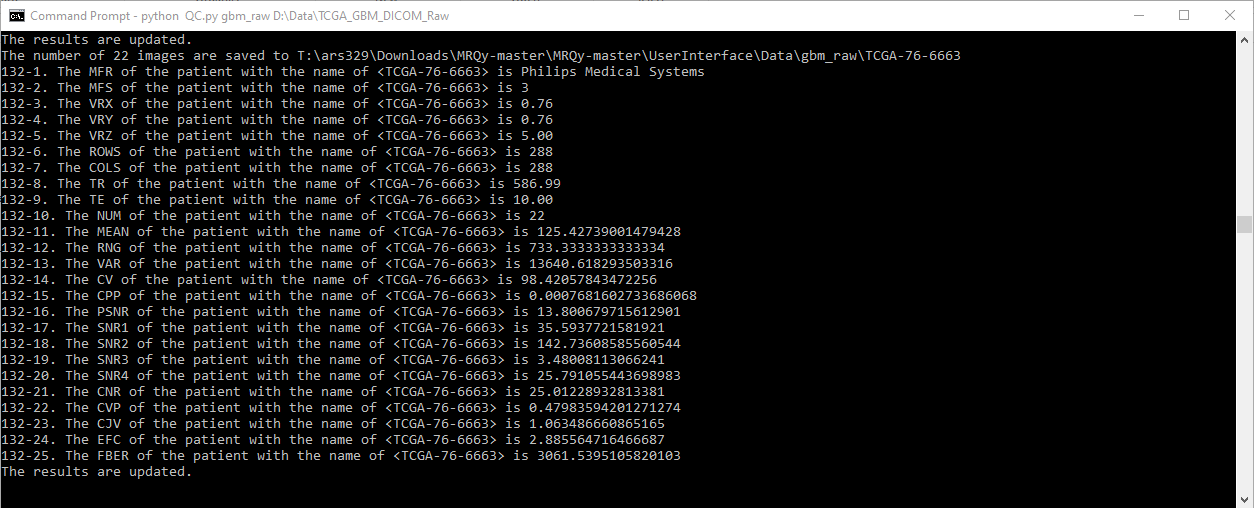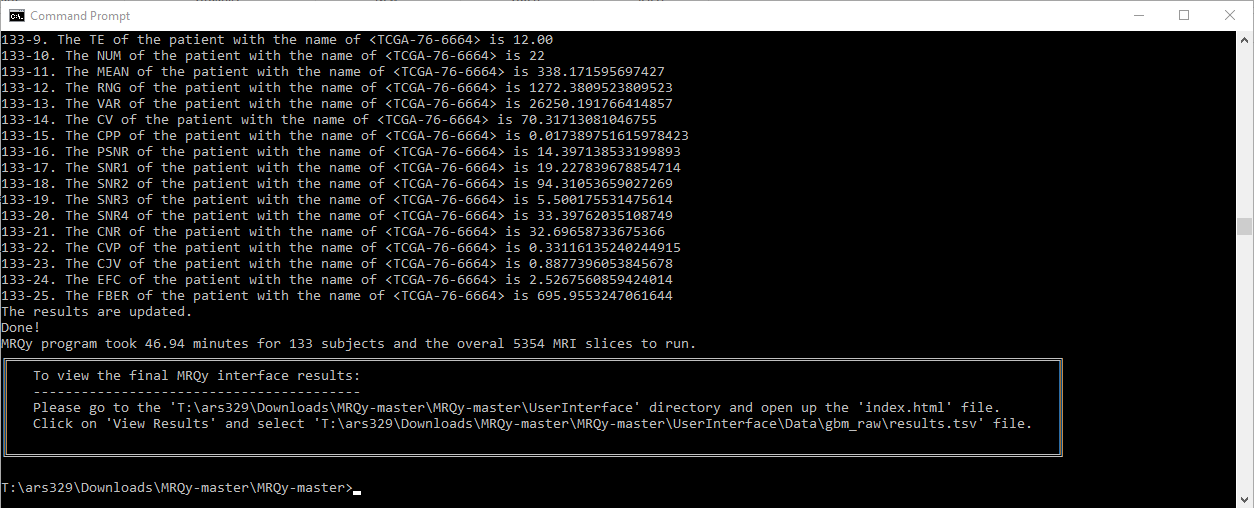
# Table of Contents
## Description
This tool takes **MRI** or **CT** datasets in the file formats as the input. \
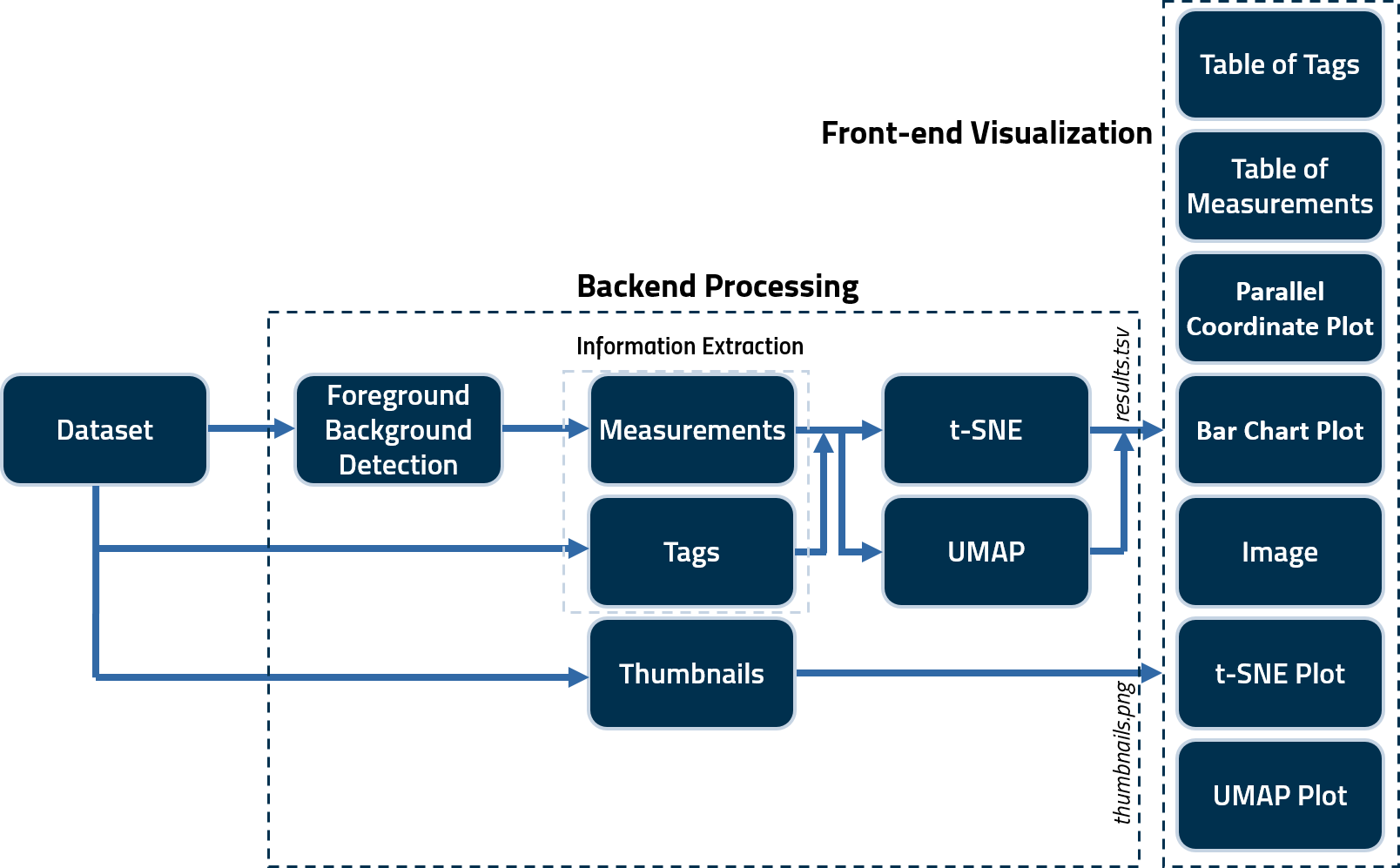
A CLI command is used to generate several tags and noise/information measurements for quality assessment. These scripts save the calculated measures in a _.tsv_ file as well as generate _.png_ thumbnails for all images in a subject volume. These are then fed to _.js_ scripts to create the user interface (_index.html_) output. A schematic illustrating the framework of the tool is as follows.
## Installation
RadQy can be installed via `pip` or `conda`.
### Using pip
```bash
pip install radqy
```
### Using conda
```bash
conda create -n radqy python=3.10
conda activate radqy
pip install radqy
```
## Running
Display the help message:
```bash
radqy --help
```
Expected Output:
```
usage: radqy [-h] [--ui-download] [--ui-run] [-s S] [-b B] [-u U] [-t {MRI,CT}]
output_folder_name inputdir [inputdir ...]
positional arguments:
output_folder_name The subfolder name in the '...\UserInterface\Data\output_folder_name' directory.
inputdir Input folder(s) containing *.dcm, *.mha, *.nii, or *.mat files.
Example: 'E:\Data\Rectal\input_data_folder'
optional arguments:
-h, --help Show this help message and exit
--ui-download Download the UserInterface.zip file
--ui-run Run the User Interface
-s S Save foreground masks (default: False)
-b B Number of samples (default: 1)
-u U Percent of middle images to process (default: 100)
-t {MRI,CT} Type of scan (MRI or CT)
```
### Running the Quality Control Script
Run the **radqy** command using the following syntax:
```bash
radqy output_folder_name "input_directory" [options]
```
Example:
```bash
radqy output_results "E:\Data\Rectal\input_data_folder" -s True -b 5 -u 50 -t CT
```
Arguments:
- **output_folder_name (required)**: The subfolder name in the `...\UserInterface\Data\output_folder_name` directory.
- **input_directory (required)**: Path to the input directory containing image files.
Options:
- **-s**: Save foreground masks (`True` or `False`). Default is `False`.
- **-b**: Number of samples. Default is `1`.
- **-u**: Percent of middle images to process. Default is `100`.
- **-t** (required): Type of scan (`MRI` or `CT`).
Notes:
- There is no need to manually create a subfolder in the Data directory; specifying its name in the command is sufficient.
- All actions will be printed in the output console for transparency.
- Thumbnail images in .png format will be saved in `...\UserInterface\Data\output_folder_name`, with each original filename as a subfolder name.
### Running the User Interface
#### Download the User Interface
To download the User Interface, run the following command:
```bash
radqy --ui-download
```
This command will download and unzip the User Interface into the appropriate directory.
#### Run the User Interface
To run the User Interface, execute:
```bash
radqy --ui-run
```
If the User Interface is not already downloaded, this command will download it automatically before launching.
#### Accessing the Front-End Interface Manually
If you prefer to access the User Interface without using the **--ui-run** command:
1. Open the Interface:
Navigate to the UserInterface directory (e.g., `C:\Users\YourUserName\.radqy\UserInterface`).
Double-click on `index.html` to open the front-end user interface.
2. Load Results:
In the interface, select the appropriate **results.tsv** file from the `...\UserInterface\Data\output_folder_name directory`.
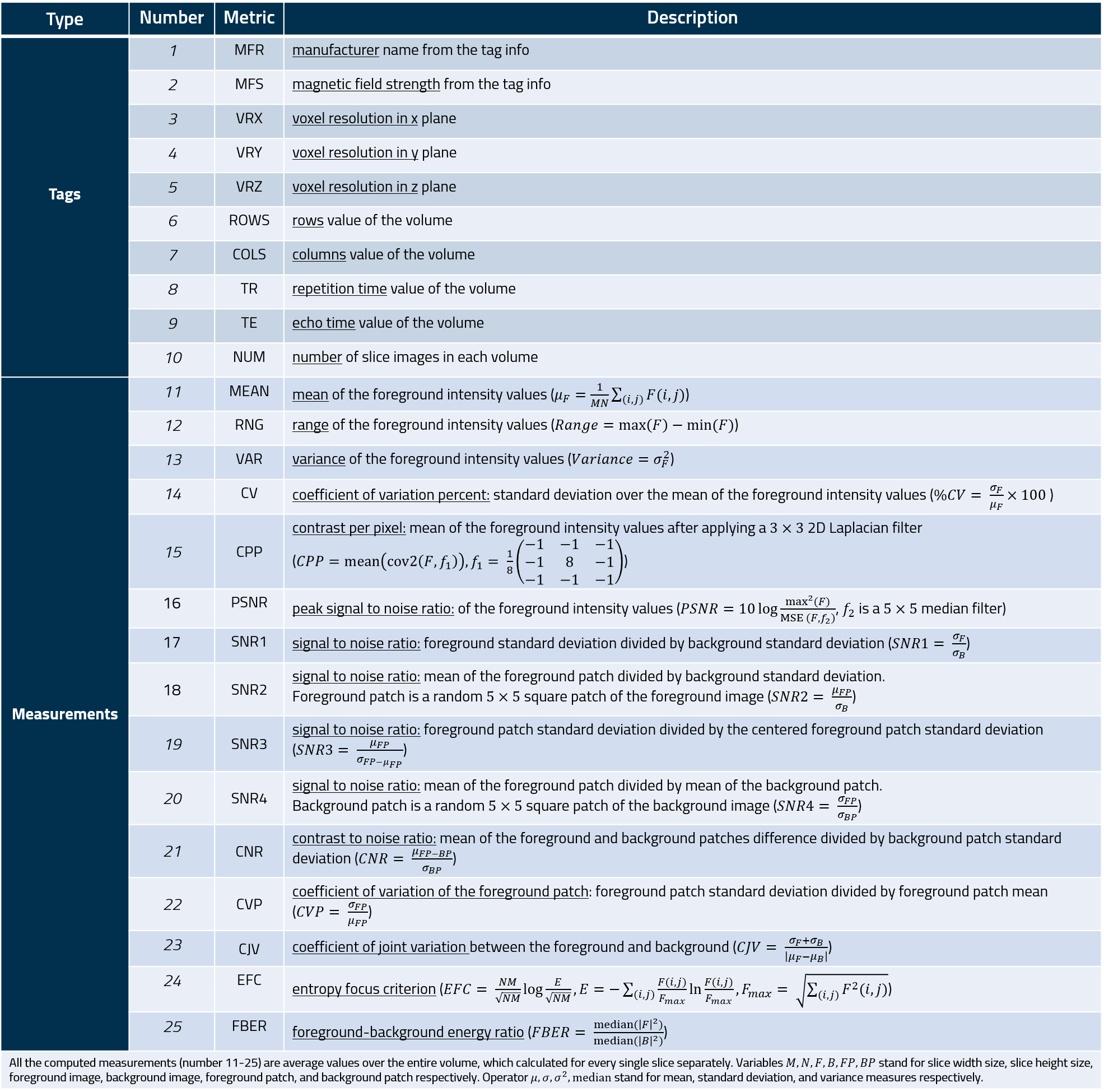
### Measurements
The measures of the MRQy tool are listed in the following table.
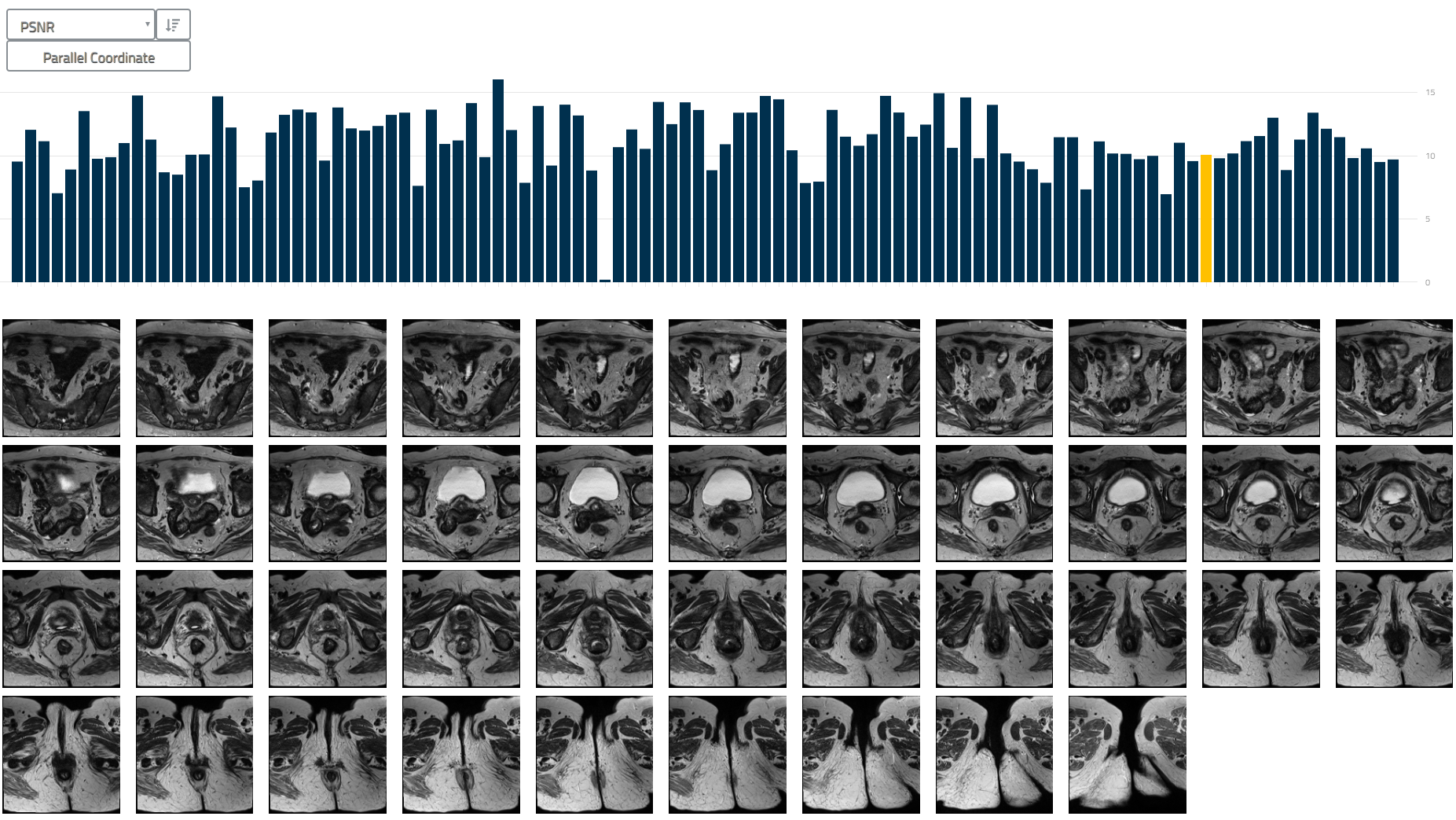
### User Interface
The following figures show the user interface of the tool (index.html).


## Feedback and usage
Please report and issues, bugfixes, ideas for enhancements via the "[Issues](https://github.com/ccipd/MRQy/issues)" tab.
Detailed usage instructions and an example of using MRQy to analyze TCIA datasets are in the [Wiki](https://github.com/ccipd/MRQy/wiki).
You can cite this in any associated publication as:
Sadri, AR, Janowczyk, A, Zou, R, Verma, R, Beig, N, Antunes, J, Madabhushi, A, Tiwari, P, Viswanath, SE, "Technical Note: MRQy — An open-source tool for quality control of MR imaging data", Med. Phys., 2020, 47: 6029-6038. https://doi.org/10.1002/mp.14593
ArXiv: https://arxiv.org/abs/2004.04871
If you do use the tool in your own work, please drop us a line to let us know.